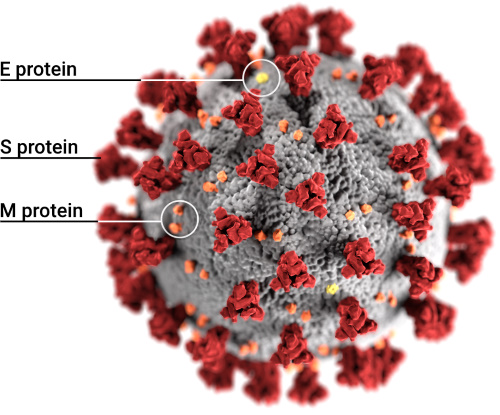
Structures
Our database currently contains information about 3783 SARS-CoV-2 protein structures and 237 additional structures of other coronaviruses. Use the filters below to select rows with attributes of interest. Next to each filter value, the number of shown/total structures from the group is displayed. Multiple values can be selected across multiple panes. To select more than one value within a single search pane, press and hold Ctrl or Shift when selecting filters. Text search can be performed using the search box on the left below the filters.
| PDB | Resol. | Released | Title | Method | Ligand IDs | Virus | Protein | Ligand category | R-work | R-free | R-merge | Synthetics | Ligand full name | Author List | Processing Software | Completeness | I/σ | Synchrotron/Beamline | Refinement Software | Deposition Date | PubMed ID | Metals | Molstack | Re-refined PDB | Re-refined MTZ | R-free after re-refinement | Re-refinement summary | Validation summary | Polymers | PQ1(PDB) | Issues |  |
Re-refined? | Raw data | Ref. | PR-free | PRSRZ | PGeometry | RNA | Re-refinement comparison | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1p9s | 2.54 Å | 2003-05-20 | X-ray | DIO | H-CoV | NSP5 (3CLpro) | Functional ligand | 19.64% | 27.89% | 14.20% | 1,4-DIETHYLENE DIOXIDE | - | 9.1 | ELETTRA (5.2R) | REFMAC | 2003-05-12 | 12746549 | - | - | - | - | N/A | - | - | Replicase polyprotein 1ab - residue 2966-3265, 3C-like proteinase | - | - | No | - | 14.7 | - | 12.7 | - | ||||||||
| 1uk4 | 2.50 Å | 2003-11-18 | X-ray | ATO | SARS-CoV | NSP5 (3CLpro) | Functional ligand | 21.30% | 23.10% | N/A | 5-mer peptide of inhibitor | CHLOROACETONE | DENZO | 97.40% | - | BSRF (3W1A) | CNS | 2003-08-14 | 14585926 | - | - | - | - | N/A | - | - | 3C-like proteinase; 5-mer peptide of inhibitor | 12.6 | - | No | - | 53.9 | 29.9 | 12.4 | - | ||||||
| 1xjr | 2.70 Å | 2005-02-01 | X-ray | - | SARS-CoV | S2M/RNA | No functional ligands | 23.14% | 24.33% | 5.60% | s2m RNA | - | MOSFLM | 99.40% | 31.3 | SSRL (BL9-1) | REFMAC | 2004-09-24 | 15630477 | MG | - | http://covid19.bioreproducibility.org/static/data/1xjr/1xjr-refined.pdb | http://covid19.bioreproducibility.org/static/data/1xjr/1xjr-refined.mtz | N/A | - | - | s2m RNA | 63.0 | - | Yes | - | 41.8 | 95.7 | 97.6 | s2m RNA | - | |||||
| 1wof | 2.00 Å | 2005-08-30 | X-ray | I12 | SARS-CoV | NSP5 (3CLpro) | Functional ligand | 20.10% | 23.90% | N/A | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]-L-ALANYL-L-VALYL-N~1~-((1S)-4-ETHOXY-4-OXO-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]-L-ALANYL-L-VALYL-N~1~-((1S)-4-ETHOXY-4-OXO-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | HKL-2000 | - | - | ROTATING ANODE () | CNS | 2004-08-18 | 16128623 | - | - | - | - | N/A | - | - | 3C-like proteinase | 19.5 | - | No | - | 45.7 | 44.2 | 23.4 | - | ||||||
| 2ajf | 2.90 Å | 2005-09-20 | X-ray | NAG | SARS-CoV | Spike | Pathogen-host interaction | 21.80% | 27.50% | N/A | beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 93.10% | 20.5 | ALS (8.2.1) | REFMAC | 2005-08-01 | 16166518 | ZN | - | http://covid19.bioreproducibility.org/static/data/2ajf/2ajf_refined.pdb | http://covid19.bioreproducibility.org/static/data/2ajf/2ajf_refined.mtz | 27.60% | Placed the coordinates in a standardized location in the unit cell. | The coordinates are NOT in a standardized location in the unit cell. | Angiotensin-converting enzyme-Related Carboxypeptidase (Ace2) - residues 19-615; SARS-coronavirus spike protein - receptor-binding domain, residues 323-502 | 4.8 | minimal | Yes | Yes | - | 16.6 | 36.9 | 15.7 | - | |||||
| 2d2d | 2.70 Å | 2005-09-20 | X-ray | ENB | SARS-CoV | NSP5 (3CLpro) | Functional ligand | 20.90% | 28.20% | N/A | ETHYL (2E,4S)-4-[((2R)-2-{[N-(TERT-BUTOXYCARBONYL)-L-VALYL]AMINO}-2-PHENYLETHANOYL)AMINO]-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENT-2-ENOATE | ETHYL (2E,4S)-4-[((2R)-2-{[N-(TERT-BUTOXYCARBONYL)-L-VALYL]AMINO}-2-PHENYLETHANOYL)AMINO]-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENT-2-ENOATE | DENZO | - | - | ROTATING ANODE () | CNS | 2005-09-08 | 16128623 | - | - | - | - | N/A | - | - | 3C-like proteinase | 15.4 | - | No | - | 13.1 | 81.0 | 9.5 | - | ||||||
| 2alv | 1.90 Å | 2006-08-08 | X-ray | CY6 | SARS-CoV | NSP5 (3CLpro) | Functional ligand | 25.50% | 31.50% | 3.80% | N-((3S,6R)-6-((S,E)-4-ETHOXYCARBONYL-1-((S)-2-OXOPYRROLIDIN-3-YL)BUT-3-EN-2-YLCARBAMOYL)-2,9-DIMETHYL-4-OXODEC-8-EN-3-YL)-5-METHYLISOXAZOLE-3-CARBOXAMIDE | N-((3S,6R)-6-((S,E)-4-ETHOXYCARBONYL-1-((S)-2-OXOPYRROLIDIN-3-YL)BUT-3-EN-2-YLCARBAMOYL)-2,9-DIMETHYL-4-OXODEC-8-EN-3-YL)-5-METHYLISOXAZOLE-3-CARBOXAMIDE | DENZO; HKL-2000 | 98.60% | 32.4 | APS (14-BM-C) | CNS | 2005-08-08 | 16250632 | - | - | http://covid19.bioreproducibility.org/static/data/2alv/2alv_refined.pdb | http://covid19.bioreproducibility.org/static/data/2alv/2alv_refined.mtz | 29.50% | Placed the coordinates in a standardized location in the unit cell. Aded link between the inhibitor and Cys145, corrected. Corrected residues Leu50, Asn51, Val125, Ile286, 301-303. C38 atom of the inhibitor, moved 5-13 oxazol moiety of the inhibitor. | Several residues and the inhibitor can be corrected. The inhibitor is not linked to the protein. The coordinates are NOT in a standardized location in the unit cell. | Replicase polyprotein 1ab - 3CL proteinase, residues 3242-3543 | 1.3 | moderate | Yes | Yes | - | 5.4 | 27.6 | 15.1 | - | |||||
| 2gx4 | 1.93 Å | 2007-05-08 | X-ray | NOL | SARS-CoV | NSP5 (3CLpro) | Functional ligand | 20.53% | 24.88% | N/A | N-[(BENZYLOXY)CARBONYL]-O-(TERT-BUTYL)-L-THREONYL-3-CYCLOHEXYL-N-[(1S)-2-HYDROXY-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}ETHYL]-L-ALANINAMIDE | N-[(BENZYLOXY)CARBONYL]-O-(TERT-BUTYL)-L-THREONYL-3-CYCLOHEXYL-N-[(1S)-2-HYDROXY-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}ETHYL]-L-ALANINAMIDE | DENZO; CrystalClear | - | - | ROTATING ANODE () | CNS | 2006-05-08 | 16884309 | - | - | - | - | N/A | - | - | 3C-like proteinase - residues 1-306 | 36.8 | - | No | - | 36.5 | 68.9 | 44.4 | - | ||||||
| 2qiq | 1.90 Å | 2008-02-12 | X-ray | CYV | SARS-CoV | NSP5 (3CLpro) | Functional ligand | 23.30% | 27.50% | N/A | ETHYL (4R)-4-{[(2R,5S)-5-{[N-(TERT-BUTOXYCARBONYL)-L-SERYL]AMINO}-6-METHYL-2-(3-METHYLBUT-2-EN-1-YL)-4-OXOHEPTANOYL]AMINO}-5-[(3R)-2-OXOPYRROLIDIN-3-YL]PENTANOATE | ETHYL (4R)-4-{[(2R,5S)-5-{[N-(TERT-BUTOXYCARBONYL)-L-SERYL]AMINO}-6-METHYL-2-(3-METHYLBUT-2-EN-1-YL)-4-OXOHEPTANOYL]AMINO}-5-[(3R)-2-OXOPYRROLIDIN-3-YL]PENTANOATE | HKL-2000 | - | - | APS (22-BM) | REFMAC | 2007-07-05 | 17855091 | - | - | http://covid19.bioreproducibility.org/static/data/2qiq/2qiq_refined.pdb | http://covid19.bioreproducibility.org/static/data/2qiq/2qiq_refined.mtz | 25.30% | Placed the coordinates in a standardized location in the unit cell. The inhibitor was re-refined after truncation to terminal carboxyl groups, assuming that the ester groups had been eliminated by hydrolysis before crystal formation, a supposition that is supported by the well-defined and abrupt limits of the electron density at the ends of both carboxyl groups. | While a large central part of the inhibitor is well defined in the electron density, the ethyl and t-butyl ester groups of the inhibitor molecule lack any electron density. The coordinates are NOT in a standardized location in the unit cell. | Replicase polyprotein 1ab - 3CL proteinase | 4.6 | significant | Yes | Yes | - | 16.6 | 17.0 | 34.5 | - | |||||
| 3d0g | 2.80 Å | 2008-07-08 | X-ray | NDG, NAG | SARS-CoV | Spike | Pathogen-host interaction | 21.40% | 27.90% | N/A | 2-acetamido-2-deoxy-alpha-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | DENZO | - | - | APS (19-BM) | REFMAC; CNS | 2008-05-01 | 18448527 | ZN | https://molstack.bioreproducibility.org/project/view/hnlY2nbqOGN6OoG7s3qz/ | http://covid19.bioreproducibility.org/static/data/3d0g/3d0g_refined.pdb | http://covid19.bioreproducibility.org/static/data/3d0g/3d0g_refined.mtz | 25.50% | Placed the coordinates in a standardized location in the unit cell. Corrected carbohydrate names, added several new carbohydrates. Fixed several rotamer outliers. | Not connected NAG molecules, not satisfactory Zn ion coordination. Unlikely assignment of the carbohydrate moieties bound to asparagine side chains (NDG rather than the expected NAG). Several carbohydrate moieties can be added to those already present, as well as to a few additional asparagine side chains. Coordinates are outside of the unit cell (not according to standardised placement) | Angiotensin-converting enzyme 2; Spike glycoprotein - residues 324-502 | 3.0 | moderate | Yes | Yes | - | 14.5 | 27.3 | 18.5 | - | ||||||
| 3d0h | 3.10 Å | 2008-07-08 | X-ray | NDG, NAG | SARS-CoV | Spike | Pathogen-host interaction | 22.10% | 30.20% | N/A | 2-acetamido-2-deoxy-alpha-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | APS (19-BM) | REFMAC | 2008-05-01 | 18448527 | ZN | https://molstack.bioreproducibility.org/project/view/WrI2XslE978LiF95PQYo/ | http://covid19.bioreproducibility.org/static/data/3d0h/3d0h_refined.pdb | http://covid19.bioreproducibility.org/static/data/3d0h/3d0h_refined.mtz | 26.60% | Placed the coordinates in a standardized location in the unit cell. Corrected carbohydrate names, added several new carbohydrates. Fixed several rotamer outliers. | Not connected NAG molecules, not satisfactory Zn ion coordination. Unlikely assignment of the carbohydrate moieties bound to asparagine side chains (NDG rather than the expected NAG). Several carbohydrate moieties can be added to those already present, as well as to a few additional asparagine side chains. Coordinates are outside of the unit cell (not according to standardised placement) | Angiotensin-converting enzyme 2; Spike glycoprotein - residues 324-502 | 11.7 | moderate | Yes | Yes | - | 6.9 | 74.0 | 12.9 | - | |||||||
| 3d0i | 2.90 Å | 2008-07-08 | X-ray | NDG, NAG | SARS-CoV | Spike | Pathogen-host interaction | 22.40% | 27.80% | N/A | 2-acetamido-2-deoxy-alpha-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | APS (19-BM) | REFMAC | 2008-05-01 | 18448527 | ZN | https://molstack.bioreproducibility.org/project/view/tyBAIB4W8gsYJT2boa8Q/ | http://covid19.bioreproducibility.org/static/data/3d0i/3d0i_refined.pdb | http://covid19.bioreproducibility.org/static/data/3d0i/3d0i_refined.mtz | 26.20% | Placed the coordinates in a standardized location in the unit cell. Corrected carbohydrate names, added several new carbohydrates. Fixed several rotamer outliers. | Not connected NAG molecules, not satisfactory Zn ion coordination. Unlikely assignment of the carbohydrate moieties bound to asparagine side chains (NDG rather than the expected NAG). Several carbohydrate moieties can be added to those already present, as well as to a few additional asparagine side chains. Coordinates are outside of the unit cell (not according to standardised placement) | Angiotensin-converting enzyme 2; Spike glycoprotein - residues 324-502 | 4.4 | moderate | Yes | Yes | - | 15.0 | 39.2 | 13.1 | - | |||||||
| 3bgf | 3.00 Å | 2008-12-02 | X-ray | - | SARS-CoV | Spike | Pathogen-host interaction | 23.40% | 28.60% | 13.20% | - | HKL-2000; DENZO | 94.40% | 6.3 | NSLS (X12C) | REFMAC | 2007-11-26 | 19324051 | - | - | - | - | N/A | - | - | Spike protein S1; F26G19 Fab; F26G19 Fab | 17.3 | - | No | - | 11.4 | 74.7 | 22.1 | - | |||||||
| 3aw0 | 2.30 Å | 2011-12-14 | X-ray | - | SARS-CoV | NSP5 (3CLpro) | Functional ligand | 22.48% | 28.64% | 8.20% | peptide ACE-SER-ALA-VAL-LEU-HIS-H | - | HKL-2000 | 99.90% | 21.786 | Photon Factory (BL-6A) | REFMAC | 2011-03-09 | 22014094 | - | - | - | - | N/A | - | - | 3C-Like Proteinase; peptide ACE-SER-ALA-VAL-LEU-HIS-H | 5.4 | - | No | - | 11.3 | 40.5 | 19.9 | - | ||||||
| 3sci | 2.90 Å | 2012-02-08 | X-ray | - | SARS-CoV | Spike | Pathogen-host interaction | 22.59% | 28.26% | N/A | - | HKL-2000 | 93.40% | - | APS (19-ID) | CNS; REFMAC | 2011-06-07 | 22291007 | ZN | - | http://covid19.bioreproducibility.org/static/data/3sci/3sci_refined_2_tls.pdb | http://covid19.bioreproducibility.org/static/data/3sci/3sci_refined_2.mtz | 27.70% | Placed the coordinates in a standardized location in the unit cell. Added a water molecule to each Zn cation and added LINKs between Zn and each of the coordinating ligands. Removed Cl ions. Fixed someRamachandran outliers. Many more Ramachandran outliers are left; the refinement is stopped due to availability of better structures for the same proteins. Sugar moieties were not added. | Zinc cations are placed without proper coordination. There are also two Cl ions in the structure, chemically feasible, but with poor/lacking electron density. Multiple Ramachandran outliers. Some residues have electron density for the sugar moieties due to glycosylation. The coordinates are NOT in a standardized location in the unit cell. | Angiotensin-converting enzyme 2 - UNP residues 19-615; Spike glycoprotein - receptor binding domain (UNP residues 306-527) | 2.4 | minimal | Yes | Yes | - | 12.9 | 29.2 | 14.0 | - | ||||||
| 3sck | 3.00 Å | 2012-02-08 | X-ray | - | SARS-CoV | Spike | Pathogen-host interaction | 23.92% | 28.53% | N/A | - | HKL-2000 | 91.70% | - | APS (19-ID) | CNS; REFMAC | 2011-06-07 | 22291007 | ZN | - | http://covid19.bioreproducibility.org/static/data/3sck/3sck_refined_tls_2.pdb | http://covid19.bioreproducibility.org/static/data/3sck/3sck_refined_tls_2.mtz | 29.50% | Placed the coordinates in a standardized location in the unit cell. Added a water molecule to each Zn cation and added LINKs between Zn and each of the coordinating ligands. Removed Cl ions. Fixed someRamachandran outliers. Many more Ramachandran outliers are left; the refinement is stopped due to availability of better structures for the same proteins. Sugar moieties were not added. | Zinc cations are placed without proper coordination. There are also two Cl ions in the structure, chemically feasible, but with poor/lacking electron density. Multiple Ramachandran outliers. Some residues have electron density for the sugar moieties due to glycosylation. The coordinates are NOT in a standardized location in the unit cell. | Angiotensin-converting enzyme 2 chimera - SEE REMARK 999; Spike glycoprotein - receptor binding domain (UNP residues 324-502) | 11.7 | minimal | Yes | Yes | - | 11.8 | 72.9 | 8.9 | - | ||||||
| 3scl | 3.00 Å | 2012-02-08 | X-ray | - | SARS-CoV | Spike | Pathogen-host interaction | 23.88% | 29.18% | N/A | - | HKL-2000 | 97.70% | - | APS (24-ID-E) | REFMAC; CNS | 2011-06-07 | 22291007 | ZN | - | http://covid19.bioreproducibility.org/static/data/3scl/3scl_refined_2_tls.pdb | http://covid19.bioreproducibility.org/static/data/3scl/3scl_refined_2_coot.mtz | 29.00% | Placed the coordinates in a standardized location in the unit cell. Added a water molecule to each Zn cation and added LINKs between Zn and each of the coordinating ligands. Replaced Cl with water molecules. Fixed multiple Ramachandran outliers. Many more Ramachandran outliers are left; the refinement is stopped due to availability of better structures for the same proteins. Sugar moieties were not added. | Zinc cations are placed without proper coordination. There are also two Cl ions in the structure, chemically feasible, but with poor/lacking electron density. Multiple Ramachandran outliers. Some residues have electron density for the sugar moieties due to glycosylation. The coordinates are NOT in a standardized location in the unit cell. | Angiotensin-converting enzyme 2 chimera - SEE REMARK 999; Spike glycoprotein - receptor binding domain (UNP residues 324-502) | 11.5 | minimal | Yes | Yes | - | 9.4 | 70.1 | 13.5 | - | ||||||
| 4rsp | 1.62 Å | 2015-06-17 | X-ray | - | MERS-CoV | NSP5 (3CLpro) | Functional ligand | 16.18% | 20.27% | N/A | Peptide inhibitor | - | MOSFLM | 95.00% | - | APS (31-ID) | PHENIX | 2014-11-10 | 26055715 | - | - | http://covid19.bioreproducibility.org/static/data/4rsp/4rsp_refined.pdb | http://covid19.bioreproducibility.org/static/data/4rsp/4rsp_refined.mtz | 21.20% | Placed the coordinates in a standardized location in the unit cell. Reset occupancies to n/m integral values. | Many residues with unbelievable occupancies (0.74/0.26, 0.61/0.39 etc.). The coordinates are NOT in a standardized location in the unit cell. | Orf1a protein; Peptide inhibitor | 79.8 | minimal | Yes | Yes | - | 79.9 | 73.8 | 88.4 | - | |||||
| 5zuv | 2.21 Å | 2019-04-10 | X-ray | - | H-CoV-229E | Spike | Functional ligand | 20.19% | 24.81% | N/A | - | HKL-3000 | 99.40% | 18.9 | SSRF (BL19U1) | REFMAC | 2018-05-08 | 30989115 | - | - | http://covid19.bioreproducibility.org/static/data/5zuv/5zuv_refined.pdb | http://covid19.bioreproducibility.org/static/data/5zuv/5zuv_refined.mtz | 23.00% | Repositioned the coordinates with ACHESYM and improved the model by adding residue Leu8 in chain c (inhibitor peptide), correcting conformations of 13 residues (Lys23b, Gln788B/A, Glu789A, Asn790B, Gln791A/B, Leu794A, Val806C, Gln839C, Thr848B, Gln856A/B), and adding 43 water molecules. | Structure of good quality, without any additional ligands and without missing side chains. Most of the protein residues outside of the reference unit cell. The fusion molecule (HR1 motif and the peptidic inhibitor) is represented in the deposit as a single protein chain with consecutive residue numbering, not as separte chains (as in 5zvk and 5zvm). | Spike glycoprotein,Spike glycoprotein,inhibitor EK1 - UNP residues 785-873 | 47.3 | moderate | Yes | Yes | - | 37.0 | 45.2 | 87.6 | - | ||||||
| 5zvk | 3.31 Å | 2019-04-10 | X-ray | - | MERS-CoV | Spike | Functional ligand | 25.90% | 31.36% | N/A | - | HKL-3000 | 99.90% | 20.7 | SSRF (BL19U1) | PHENIX | 2018-05-11 | 30989115 | - | - | - | - | N/A | - | Structure of good quality, without any additional ligands and with no missing side chains. Most of the protein residues outside of the reference unit cell. | Human Coronavirus MERS HR1 motif - UNP residues 984-1062; pan-CoV inhibitory peptide EK1 | 7.4 | minimal | No | - | 5.6 | 26.1 | 47.8 | - | |||||||
| 5zvm | 3.30 Å | 2019-04-10 | X-ray | - | SARS-CoV | Spike | Functional ligand | 24.76% | 30.14% | N/A | - | HKL-3000 | 99.90% | 11.9 | SSRF (BL19U1) | REFMAC | 2018-05-11 | 30989115 | - | - | - | - | N/A | not done | The coordinates are NOT in a standardized location in the unit cell. | Spike glycoprotein - UNP residues 892-970; pan-CoV inhibitory peptide EK1 | 13.4 | minimal | No | - | 7.0 | 16.9 | 74.5 | - | |||||||
| 6u7e | 3.00 Å | 2019-11-13 | X-ray | NAG | H-CoV-229E | Spike | Pathogen-host interaction | 20.35% | 24.39% | 7.10% | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.80% | 11.06 | CLSI (08ID-1) | PHENIX | 2019-09-02 | 31650956 | ZN | - | - | - | N/A | - | - | Aminopeptidase N - ectodomain (UNP residues 66-967); Spike glycoprotein | 65.3 | - | No | 41.3 | 86.6 | 77.5 | - | ||||||||
| 6u7f | 2.75 Å | 2019-11-13 | X-ray | NAG | H-CoV-229E | Spike | Pathogen-host interaction | 19.04% | 22.51% | 6.80% | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.80% | 16.24 | CLSI (08ID-1) | PHENIX | 2019-09-02 | 31650956 | ZN | - | - | - | N/A | - | - | Aminopeptidase N - ectodomain (UNP residues 66-967); Spike glycoprotein | 76.1 | - | No | 60.0 | 86.4 | 84.4 | - | ||||||||
| 6u7g | 2.35 Å | 2019-11-13 | X-ray | NAG | H-CoV-229E | Spike | Pathogen-host interaction | 17.86% | 21.84% | 5.20% | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.70% | 14.9 | APS (23-ID-B) | PHENIX | 2019-09-02 | 31650956 | ZN | - | http://covid19.bioreproducibility.org/static/data/6u7g/6u7g_refined.pdb | http://covid19.bioreproducibility.org/static/data/6u7g/6u7g_refined.mtz | 22.40% | Fixed minor issues with geometry and solvent | - | Aminopeptidase N - ectodomain (UNP residues 66-967); Spike protein - receptor-binding domain (UNP residues 292-432) | 55.4 | minimal | Yes | 66.6 | 35.5 | 83.3 | - | ||||||||
| 6u7h | 3.10 Å | 2019-11-13 | Cryo-EM | NAG | H-CoV-229E | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2019-09-02 | 31650956 | - | - | - | - | N/A | - | - | spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6jx7 | 3.31 Å | 2020-01-15 | Cryo-EM | MAN, NAG | FIPV | Spike | No functional ligands | N/A | N/A | N/A | alpha-D-mannopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2019-04-22 | 31900356 | - | - | - | - | N/A | - | - | Feline Infectious Peritonitis Virus Spike Protein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6noz | 1.95 Å | 2020-01-22 | X-ray | - | PEDV | NSP3: PLpro | No functional ligands | 14.47% | 19.31% | 16.40% | - | HKL-2000 | 100.00% | 6.4 | APS (31-ID) | PHENIX | 2019-01-16 | - | ZN | - | - | - | N/A | - | - | Polyprotein - UNP Residues 1688-1933 | 85.4 | - | No | - | - | 86.1 | 92.3 | 89.9 | - | ||||||
| 6lu7 | 2.16 Å | 2020-02-05 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.20% | 23.50% | 18.90% | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | - | xia2 | 100.00% | 6.3 | SSRF (BL17U1) | PHENIX | 2020-01-26 | 32272481 | - | https://molstack.bioreproducibility.org/project/view/x9jJQnlGN25TRBSRdxJm/ | http://covid19.bioreproducibility.org/static/data/6lu7/6lu7_refined.pdb | http://covid19.bioreproducibility.org/static/data/6lu7/6lu7_refined.mtz | 22.50% | The phenyl group was removed from the inhibitor. After re-refinement of the model negative density disappeared. A few minor model corrections. | The terminal phenyl group of the inhibitor with negative difference electron density, possible hydrolysis. | main protease - 3C-like proteinase; N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | 35.0 | moderate | Yes | Yes | - | 49.8 | 29.9 | 66.4 | - | |||||
| 6lvn | 2.47 Å | 2020-02-26 | X-ray | - | SARS-CoV-2 | Spike | No functional ligands | 21.48% | 25.80% | 13.24% | - | XDS | 99.22% | 19.06 | ROTATING ANODE () | PHENIX | 2020-02-04 | - | - | - | http://covid19.bioreproducibility.org/static/data/6lvn/6lvn_refined_tls.pdb | http://covid19.bioreproducibility.org/static/data/6lvn/6lvn_refined.mtz | 26.90% | Brought all four chains in one bundle. Modified residues: 18, 30, 2, 29. Added 34water molecules. | Structure of good quality. No ligands bound. No Ramachandran outliers, no rotamer outliers, and no missing side chains. Four helices are not placed in the ASU as a single oligomer, but as two independent dimers | Spike protein S2 - HR2 domain | 49.1 | minimal | Yes | Yes | - | - | 28.1 | 76.3 | 69.0 | - | |||||
| 6lxt | 2.90 Å | 2020-02-26 | X-ray | PG4 | SARS-CoV-2 | Spike | No functional ligands | 25.93% | 28.96% | 16.00% | TETRAETHYLENE GLYCOL | MOSFLM | 94.02% | 6.3 | ROTATING ANODE () | REFMAC | 2020-02-11 | 32231345 | ZN | - | - | - | N/A | not done | Structure of good quality. It has one Ramachandran outlier, probably justified. No missing side-chains. Several side-chain outliers, but most are not trivial to fix - a common problem at low resolution (% outliers is low). Most of the protein is outside of the unit cell. A metal cation (Zn2+) is missing between Asp1199 of chain B and Asp1199 of chain F. At the interface between Asp1163 and Asp1165 of chain B and a symmetry mate of Glu1188 and Asp1184 of chain A (crystallographic interface between two six-helix bundles), a molecule of PEG is modeled instead of, most probably, one or two Zn2+ ions. This PEG molecule is hilariously mismodelled: it has severe clashes with the protein residues, all contacts are polar-to-nonpolar, and it is too curled-up | Spike protein S2, Spike protein S2 - HR1 domain,HR2 domain | 10.6 | moderate | No | - | 10.1 | 49.9 | 30.3 | - | |||||||
| 6vsb | 3.46 Å | 2020-02-26 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-02-10 | 32075877 | - | - | - | - | N/A | - | - | SARS-CoV-2 spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6vv5 | 3.50 Å | 2020-02-26 | Cryo-EM | NAG, PAM | PEDV | Spike | No functional ligands | N/A | N/A | N/A | PALMITOLEIC ACID | 2-acetamido-2-deoxy-beta-D-glucopyranose; PALMITOLEIC ACID | - | - | () | PHENIX | 2020-02-17 | 33378641 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 6vsj | 3.94 Å | 2020-03-04 | Cryo-EM | NAG | Murine-CoV-A59 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-02-11 | 32150576 | - | - | - | - | N/A | - | - | Spike glycoprotein; Carcinoembryonic antigen-related cell adhesion molecule 1 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6vw1 | 2.68 Å | 2020-03-04 | X-ray | NAG | SARS-CoV/SARS-CoV-2 | Spike | Pathogen-host interaction | 19.70% | 22.88% | 8.10% | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 96.70% | 12.08 | APS (24-ID-E) | PHENIX; REFMAC | 2020-02-18 | 32225175 | ZN | - | http://covid19.bioreproducibility.org/static/data/6vw1/6vw1_refined.pdb | http://covid19.bioreproducibility.org/static/data/6vw1/6vw1_refined.mtz | 24.00% | Placed the coordinates in a standardized location in the unit cell. MC - refined the structure | Zn2+ cations have abnormally high B factors (135 and 196 Å2) and do not make any contacts with their environment that would indicate proper coordination. Otherwise the structure seems to be in good agreement with the electron density. The coordinates are NOT in a standardized location in the unit cell. | Angiotensin-converting enzyme 2; SARS-CoV-2 chimeric RBD | 52.0 | minimal | Yes | Yes | - | 56.3 | 76.4 | 46.3 | http://covid19.bioreproducibility.org/static/data/6vw1/6vw1_refinement_report.pdf | ||||||
| 6vww | 2.20 Å | 2020-03-04 | X-ray | ACY | SARS-CoV-2 | NSP15 | No functional ligands | 15.77% | 17.77% | 17.40% | ACETIC ACID | HKL-3000 | 98.50% | 10.0 | APS (19-ID) | PHENIX | 2020-02-20 | 32304108 | MG | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 85.8 | - | No | 93.0 | 95.3 | 82.9 | - | ||||||||
| 6vxs | 2.03 Å | 2020-03-04 | X-ray | NHE | SARS-CoV-2 | NSP3: Macro | No functional ligands | 18.64% | 23.35% | N/A | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID | HKL-3000 | 90.80% | 8.44 | APS (19-ID) | PHENIX | 2020-02-24 | 32939273 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 74.3 | - | No | 51.6 | 89.6 | 85.0 | - | |||||||
| 6y2e | 1.75 Å | 2020-03-04 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 17.12% | 22.24% | 8.20% | - | XDS | 100.00% | 15.0 | BESSY (14.2) | REFMAC | 2020-02-15 | 32198291 | - | - | http://covid19.bioreproducibility.org/static/data/6y2e/6y2e_refined.pdb | http://covid19.bioreproducibility.org/static/data/6y2e/6y2e_refined.mtz | 22.06% | Fixed minor issues with side chain rotamers. | Protein chains are modeled well. | Replicase polyprotein 1ab | 62.6 | minimal | Yes | Yes | - | 62.8 | 64.4 | 72.7 | - | ||||||
| 6y2f | 1.95 Å | 2020-03-04 | X-ray | O6K | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.79% | 21.91% | 7.60% | ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | XDS | 100.00% | 15.5 | BESSY (14.2) | REFMAC | 2020-02-15 | 32198291 | - | - | http://covid19.bioreproducibility.org/static/data/6y2f/6y2f_refined.pdb | http://covid19.bioreproducibility.org/static/data/6y2f/6y2f_refined.mtz | 20.60% | Re-refinement with a sodium cation modeled at the highest peak site. A few side chain rotamer corrections. Application of TLS parameters. | Structure of good quality. High positive peak coordinated by five carbonyl oxygen atoms and one water molecule; octahedral coordination. | Replicase polyprotein 1ab | 42.4 | moderate | Yes | Yes | - | 65.8 | 37.2 | 57.5 | - | |||||
| 6y2g | 2.20 Å | 2020-03-04 | X-ray | O6K | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.62% | 24.73% | 17.00% | ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | XDS | 100.00% | 13.1 | BESSY (14.2) | REFMAC | 2020-02-15 | 32198291 | - | - | http://covid19.bioreproducibility.org/static/data/6y2g/6y2g_refined.pdb | http://covid19.bioreproducibility.org/static/data/6y2g/6y2g_refined.mtz | 24.00% | Fixed minor issues with side chain rotamers and a couple of orphaned solvent molecules. | Protein chains are modeled well. | Replicase polyprotein 1ab | 46.9 | minimal | No | Yes | - | 37.8 | 78.5 | 52.9 | - | |||||
| 5r7y | 1.65 Å | 2020-03-11 | X-ray | JFM | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.94% | 23.67% | 11.30% | N-(2-phenylethyl)methanesulfonamide | N-(2-phenylethyl)methanesulfonamide | XDS | 99.40% | 5.8 | Diamond (I04-1) | REFMAC | 2020-03-03 | 33028810 | - | - | - | - | N/A | not done | Weak density for the ligand on the primary 2Fo-Fc map. | 3C-like proteinase | - | moderate | No | 48.3 | - | 51.2 | - | |||||||
| 5r7z | 1.59 Å | 2020-03-11 | X-ray | HWH | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.95% | 23.30% | 9.60% | ~{N}-[2-(5-fluoranyl-1~{H}-indol-3-yl)ethyl]ethanamide | ~{N}-[2-(5-fluoranyl-1~{H}-indol-3-yl)ethyl]ethanamide | XDS | 99.30% | 6.2 | Diamond (I04-1) | REFMAC | 2020-03-03 | 33028810 | - | https://molstack.bioreproducibility.org/collection/view/GqjIgzdHOk3yuQaO0ukE/ | - | - | 24.30% | not done | Weak density for the ligand on the primary 2Fo-Fc map. | 3C-like proteinase | 40.9 | moderate | No | 51.9 | 42.5 | 63.3 | - | |||||||
| 5r80 | 1.93 Å | 2020-03-11 | X-ray | RZG | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.30% | 23.46% | 11.30% | methyl 4-sulfamoylbenzoate | methyl 4-sulfamoylbenzoate | XDS | 98.60% | 6.0 | Diamond (I04-1) | REFMAC | 2020-03-03 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 24.5 | - | No | 50.4 | 33.0 | 41.0 | - | |||||||
| 5r81 | 1.95 Å | 2020-03-11 | X-ray | RZJ | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.20% | 24.94% | 16.20% | 1-methyl-3,4-dihydro-2~{H}-quinoline-7-sulfonamide | 1-methyl-3,4-dihydro-2~{H}-quinoline-7-sulfonamide | XDS | 99.90% | 4.0 | Diamond (I04-1) | REFMAC | 2020-03-03 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 37.2 | - | No | 35.7 | 64.3 | 50.5 | - | |||||||
| 5r82 | 1.31 Å | 2020-03-11 | X-ray | RZS | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.03% | 21.16% | 5.00% | 6-(ethylamino)pyridine-3-carbonitrile | 6-(ethylamino)pyridine-3-carbonitrile | XDS | 88.70% | 9.7 | Diamond (I04-1) | REFMAC | 2020-03-03 | 33028810 | - | - | - | - | 21.20% | not done | Weak density for the ligand on the primary 2Fo-Fc map. | 3C-like proteinase | - | moderate | No | 72.7 | - | 50.0 | - | |||||||
| 5r83 | 1.58 Å | 2020-03-11 | X-ray | K0G | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.58% | 21.49% | 10.00% | N-phenyl-N'-pyridin-3-ylurea | N-phenyl-N'-pyridin-3-ylurea | XDS | 99.60% | 6.7 | Diamond (I04-1) | REFMAC | 2020-03-03 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 58.6 | - | No | 70.0 | 59.9 | 61.8 | - | |||||||
| 5r84 | 1.83 Å | 2020-03-11 | X-ray | GWS | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.67% | 29.26% | 14.00% | 2-cyclohexyl-~{N}-pyridin-3-yl-ethanamide | 2-cyclohexyl-~{N}-pyridin-3-yl-ethanamide | XDS | 99.70% | 5.6 | Diamond (I04-1) | REFMAC | 2020-03-03 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 12.7 | - | No | 9.2 | 45.4 | 41.8 | - | |||||||
| 6m03 | 2.00 Å | 2020-03-11 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 19.62% | 24.58% | 3.90% | - | XDS | 99.50% | 16.29 | NFPSS (BL18U) | PHENIX | 2020-02-19 | 35380892 | - | - | http://covid19.bioreproducibility.org/static/data/6m03/6m03_refined.pdb | http://covid19.bioreproducibility.org/static/data/6m03/6m03_refined.mtz | 24.21% | Placed the coordinates in a standardized location in the unit cell. Added TLS (9 groups), added waters, fixed ramachandran outliers, Flipped side chains GLN 74, HIS 164, ASN 203. Changed rotamers MET 17, MET 165, ARG 298, GLN 189. Added alternative conformation for PHE 294. | Flipped sidechains, missing rotamers. The coordinates are NOT in a standardized location in the unit cell. No ligands, no metals. | main protease | 51.7 | moderate | Yes | Yes | - | 39.5 | 68.9 | 70.0 | - | ||||||
| 6m17 | 2.90 Å | 2020-03-11 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; LEUCINE | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-02-24 | 32132184 | ZN | - | - | - | N/A | - | Poor map | Sodium-dependent neutral amino acid transporter B(0)AT1; Angiotensin-converting enzyme 2; SARS-coV-2 Receptor Binding Domain | N/A | moderate | No | - | N/A | N/A | N/A | - | ||||||||
| 6vxx | 2.80 Å | 2020-03-11 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-02-25 | 32155444 | - | - | - | - | N/A | - | - | SARS-CoV-2 spike glycoprotein - ectodomain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6vyb | 3.20 Å | 2020-03-11 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-02-25 | 32155444 | - | - | - | - | N/A | - | - | Spike glycoprotein - ectodomain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6vyo | 1.70 Å | 2020-03-11 | X-ray | MES | SARS-CoV-2 | Nucleocapsid | No functional ligands | 15.92% | 20.53% | 13.10% | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | HKL-3000 | 99.70% | 6.9 | APS (19-ID) | PHENIX | 2020-02-27 | - | ZN | - | http://covid19.bioreproducibility.org/static/data/6vyo/6vyo_refined.pdb | http://covid19.bioreproducibility.org/static/data/6vyo/6vyo_refined.mtz | 21.20% | - | - | Nucleoprotein - RNA binding domain | 76.5 | - | Yes | - | 77.9 | 68.4 | 85.6 | - | ||||||
| 6w01 | 1.90 Å | 2020-03-11 | X-ray | - | SARS-CoV-2 | NSP15 | No functional ligands | 16.05% | 18.49% | 11.20% | - | HKL-3000 | 99.30% | 13.11 | APS (19-ID) | PHENIX | 2020-02-28 | 32304108 | - | - | - | - | N/A | z | - | Uridylate-specific endoribonuclease | 80.8 | - | No | 90.3 | 72.3 | 82.9 | - | ||||||||
| 6w02 | 1.50 Å | 2020-03-11 | X-ray | APR | SARS-CoV-2 | NSP3: Macro | Functional ligand | 14.96% | 17.29% | 7.50% | ADENOSINE-5-DIPHOSPHORIBOSE | ADENOSINE-5-DIPHOSPHORIBOSE | HKL-3000 | 80.80% | 25.3 | APS (19-ID) | PHENIX | 2020-02-28 | 32939273 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 85.3 | - | No | 94.4 | 89.6 | 83.5 | - | |||||||
| 6y84 | 1.39 Å | 2020-03-11 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 17.79% | 20.00% | 5.90% | - | xia2; DIALS | 97.50% | 9.7 | Diamond (I04-1) | BUSTER | 2020-03-03 | - | - | - | http://covid19.bioreproducibility.org/static/data/6y84/6y84_refined_1.pdb | http://covid19.bioreproducibility.org/static/data/6y84/6y84_refined_1.mtz | 18.60% | - | Structure of good quality. | Replicase polyprotein 1ab | 64.6 | minimal | Yes | - | - | 81.6 | 39.8 | 82.6 | - | ||||||
| 6lzg | 2.50 Å | 2020-03-18 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 18.84% | 21.56% | 12.90% | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 100.00% | 26.7 | SSRF (BL17U1) | PHENIX | 2020-02-19 | 32275855 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | 41.1 | - | No | - | 69.3 | 33.2 | 55.6 | - | |||||||
| 6m0j | 2.45 Å | 2020-03-18 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 19.20% | 22.69% | 12.00% | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 100.00% | 24.2 | SSRF (BL17U) | PHENIX | 2020-02-21 | 32225176 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike receptor binding domain | 52.4 | - | No | - | 58.4 | 59.6 | 61.8 | - | |||||||
| 6m3m | 2.70 Å | 2020-03-18 | X-ray | - | SARS-CoV-2 | Nucleocapsid | No functional ligands | 25.78% | 29.34% | 10.43% | - | CrysalisPro | 99.50% | 14.97 | ROTATING ANODE () | PHENIX | 2020-03-04 | 32363136 | - | - | - | - | 28.90% | not done | Structure of good quality. No ligands. Some ramachandran outliers at the C-terminus. Many slashed side chains due to the lack of electron density – some can be built. | SARS-CoV-2 nucleocapsid protein - N-terminal RNA binding domain | 4.4 | minimal | No | - | 9.0 | 25.2 | 33.0 | - | |||||||
| 6w4b | 2.95 Å | 2020-03-18 | X-ray | - | SARS-CoV-2 | NSP9 | No functional ligands | 23.98% | 27.60% | 19.00% | - | HKL-3000 | 97.10% | 17.4 | APS (19-BM) | PHENIX | 2020-03-10 | - | - | - | - | - | N/A | - | - | Non-structural protein 9 | 6.6 | - | No | - | 16.1 | 24.0 | 36.6 | - | |||||||
| 6w4h | 1.80 Å | 2020-03-18 | X-ray | SO3, SAM, BDF | SARS-CoV-2 | NSP10/NSP16 | Protein-protein complex | 14.90% | 16.27% | 6.00% | S-ADENOSYLMETHIONINE | SULFITE ION; S-ADENOSYLMETHIONINE; beta-D-fructopyranose | HKL-3000 | 100.00% | 29.3 | APS (21-ID-F) | REFMAC | 2020-03-10 | 32994211 | ZN | - | - | - | N/A | - | - | SARS-CoV-2 NSP16 - UNP residues 6799-7096; SARS-CoV-2 NSP10 - UNP residues 4254-4392 | 62.5 | - | No | 96.6 | 16.1 | 87.0 | - | |||||||
| 6y7m | 1.90 Å | 2020-03-18 | X-ray | OEW | SARS-CoV | NSP5 (3CLpro) | Functional ligand | 20.62% | 25.76% | 4.80% | ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclohexyl-1-[[(2~{S},3~{R})-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclohexyl-1-[[(2~{S},3~{R})-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | XDS | 99.40% | 12.8 | PETRA III, DESY (P11) | REFMAC | 2020-03-01 | 32198291 | - | - | http://covid19.bioreproducibility.org/static/data/6y7m/6y7m_refined.pdb | http://covid19.bioreproducibility.org/static/data/6y7m/6y7m_refined.mtz | 25.60% | Added one water molecule. Some minor difficult to correction issues. | Structure of good quality. | Replicase polyprotein 1a | 29.2 | minimal | Yes | Yes | - | 28.6 | 45.7 | 60.0 | - | |||||
| 5re4 | 1.88 Å | 2020-03-25 | X-ray | SZY | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.91% | 26.64% | 22.80% | N-(4-methylpyridin-3-yl)acetamide | N-(4-methylpyridin-3-yl)acetamide | XDS | 99.50% | 3.3 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 30.1 | - | No | 22.0 | 59.9 | 54.3 | - | |||||||
| 5re5 | 2.07 Å | 2020-03-25 | X-ray | T0J | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.11% | 26.49% | 25.90% | N~1~-phenylpiperidine-1,4-dicarboxamide | N~1~-phenylpiperidine-1,4-dicarboxamide | XDS | 99.90% | 4.1 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 36.4 | - | No | 23.2 | 73.6 | 52.2 | - | |||||||
| 5re6 | 1.87 Å | 2020-03-25 | X-ray | O0S | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.57% | 25.05% | 23.00% | N-{4-[(pyrimidin-2-yl)oxy]phenyl}acetamide | N-{4-[(pyrimidin-2-yl)oxy]phenyl}acetamide | XDS | 99.80% | 4.3 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 46.0 | - | No | 34.8 | 64.3 | 68.4 | - | |||||||
| 5re7 | 1.79 Å | 2020-03-25 | X-ray | T0S | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.73% | 22.50% | 12.60% | N-[(4-sulfamoylphenyl)methyl]acetamide | N-[(4-sulfamoylphenyl)methyl]acetamide | XDS | 99.40% | 6.1 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 23.9 | - | No | 60.1 | 22.9 | 40.1 | - | |||||||
| 5re8 | 1.81 Å | 2020-03-25 | X-ray | T0V | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.03% | 24.83% | 17.10% | 1-(3-fluorophenyl)-N-[(furan-2-yl)methyl]methanamine | 1-(3-fluorophenyl)-N-[(furan-2-yl)methyl]methanamine | XDS | 99.80% | 4.6 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 34.5 | - | No | 36.8 | 52.1 | 56.2 | - | |||||||
| 5re9 | 1.72 Å | 2020-03-25 | X-ray | LPZ | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.72% | 22.48% | 8.20% | 2-(4-methylphenoxy)-1-(4-methylpiperazin-4-ium-1-yl)ethanone | 2-(4-methylphenoxy)-1-(4-methylpiperazin-4-ium-1-yl)ethanone | XDS | 98.70% | 8.0 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 18.0 | - | No | 60.5 | 13.3 | 35.9 | - | |||||||
| 5rea | 1.63 Å | 2020-03-25 | X-ray | JGP | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.52% | 22.82% | 9.60% | (azepan-1-yl)(2H-1,3-benzodioxol-5-yl)methanone | (azepan-1-yl)(2H-1,3-benzodioxol-5-yl)methanone | XDS | 99.50% | 6.2 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | 56.8 | - | 48.4 | - | |||||||
| 5reb | 1.68 Å | 2020-03-25 | X-ray | T0Y | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.55% | 22.37% | 9.30% | 1-[(thiophen-3-yl)methyl]piperidin-4-ol | 1-[(thiophen-3-yl)methyl]piperidin-4-ol | XDS | 99.50% | 6.9 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 40.4 | - | No | 61.6 | 29.3 | 65.7 | - | |||||||
| 5rec | 1.73 Å | 2020-03-25 | X-ray | T1J | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.26% | 23.71% | 12.10% | 2-{[(1H-benzimidazol-2-yl)amino]methyl}phenol | 2-{[(1H-benzimidazol-2-yl)amino]methyl}phenol | XDS | 99.60% | 5.9 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 49.4 | - | No | 47.7 | 64.3 | 61.8 | - | |||||||
| 5red | 1.47 Å | 2020-03-25 | X-ray | JJG | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.14% | 21.12% | 8.50% | 4-[2-(phenylsulfanyl)ethyl]morpholine | 4-[2-(phenylsulfanyl)ethyl]morpholine | XDS | 98.40% | 6.2 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 34.1 | - | No | 73.0 | 17.6 | 53.7 | - | |||||||
| 5ree | 1.77 Å | 2020-03-25 | X-ray | T1M | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.32% | 24.24% | 17.00% | (2R,3R)-1-benzyl-2-methylpiperidin-3-ol | (2R,3R)-1-benzyl-2-methylpiperidin-3-ol | XDS | 99.80% | 4.5 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 44.3 | - | No | 42.6 | 64.3 | 57.3 | - | |||||||
| 5ref | 1.61 Å | 2020-03-25 | X-ray | 6SU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.81% | 24.61% | 12.10% | methyl 3-(methylsulfonylamino)benzoate | methyl 3-(methylsulfonylamino)benzoate | XDS | 99.10% | 4.7 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 38.9 | - | No | 38.9 | 55.9 | 59.0 | - | |||||||
| 5reg | 1.67 Å | 2020-03-25 | X-ray | LWA | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.92% | 22.72% | 11.90% | (2~{S})-~{N}-(4-aminocarbonylphenyl)oxolane-2-carboxamide | (2~{S})-~{N}-(4-aminocarbonylphenyl)oxolane-2-carboxamide | XDS | 97.20% | 5.2 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 21.4 | - | No | 57.9 | 21.0 | 38.8 | - | |||||||
| 5reh | 1.80 Å | 2020-03-25 | X-ray | AWP | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.71% | 25.73% | 18.00% | 1-cyclohexyl-3-(2-pyridin-4-ylethyl)urea | 1-cyclohexyl-3-(2-pyridin-4-ylethyl)urea | XDS | 98.70% | 5.4 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 26.4 | - | No | 28.8 | 59.9 | 39.6 | - | |||||||
| 5rei | 1.82 Å | 2020-03-25 | X-ray | T1S | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.32% | 24.01% | 16.90% | 4-[(3-chlorophenyl)methyl]morpholine | 4-[(3-chlorophenyl)methyl]morpholine | XDS | 99.80% | 5.3 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 53.1 | - | No | 44.7 | 68.7 | 67.6 | - | |||||||
| 5rej | 1.72 Å | 2020-03-25 | X-ray | T1V | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.00% | 24.05% | 18.20% | 1-{4-[(thiophen-2-yl)sulfonyl]piperazin-1-yl}ethan-1-one | 1-{4-[(thiophen-2-yl)sulfonyl]piperazin-1-yl}ethan-1-one | XDS | 99.50% | 4.0 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 41.0 | - | No | 44.4 | 52.1 | 61.4 | - | |||||||
| 5rek | 1.74 Å | 2020-03-25 | X-ray | T1Y | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.27% | 23.04% | 17.60% | 1-{4-[(3-fluorophenyl)sulfonyl]piperazin-1-yl}ethan-1-one | 1-{4-[(3-fluorophenyl)sulfonyl]piperazin-1-yl}ethan-1-one | XDS | 99.20% | 3.9 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 53.7 | - | No | 54.6 | 55.9 | 71.8 | - | |||||||
| 5rel | 1.62 Å | 2020-03-25 | X-ray | T2G | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.21% | 21.97% | 10.90% | 1-{4-[(3-methylphenyl)methyl]piperazin-1-yl}ethan-1-one | 1-{4-[(3-methylphenyl)methyl]piperazin-1-yl}ethan-1-one | XDS | 99.70% | 5.6 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 53.7 | - | No | 65.4 | 52.1 | 64.7 | - | |||||||
| 5rem | 1.96 Å | 2020-03-25 | X-ray | T2J | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.50% | 24.64% | 16.40% | 1 1-(4-(2-nitrophenyl)piperazin-1-yl)ethan-1-one | 1 1-(4-(2-nitrophenyl)piperazin-1-yl)ethan-1-one | XDS | 99.80% | 4.7 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 38.9 | - | No | 38.7 | 59.9 | 55.2 | - | |||||||
| 5ren | 2.15 Å | 2020-03-25 | X-ray | T2V | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.38% | 27.79% | 27.70% | 1-[(3R)-3-(1,3-benzothiazol-2-yl)piperidin-1-yl]ethan-1-one | 1-[(3R)-3-(1,3-benzothiazol-2-yl)piperidin-1-yl]ethan-1-one | XDS | 99.20% | 3.6 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 19.4 | - | No | 15.2 | 48.6 | 49.2 | - | |||||||
| 5reo | 1.88 Å | 2020-03-25 | X-ray | T2Y | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.76% | 22.68% | 21.10% | N-[(2H-1,3-benzodioxol-5-yl)methyl]acetamide | N-[(2H-1,3-benzodioxol-5-yl)methyl]acetamide | XDS | 99.60% | 4.5 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 53.1 | - | No | 58.4 | 68.7 | 53.9 | - | |||||||
| 5rep | 1.81 Å | 2020-03-25 | X-ray | T3G | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.83% | 23.11% | 15.80% | 1-{4-[(2,6-difluorophenyl)sulfonyl]piperazin-1-yl}ethan-1-one | 1-{4-[(2,6-difluorophenyl)sulfonyl]piperazin-1-yl}ethan-1-one | XDS | 99.70% | 5.3 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 63.1 | - | No | 53.9 | 78.5 | 68.6 | - | |||||||
| 5rer | 1.88 Å | 2020-03-25 | X-ray | T3J | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.72% | 25.26% | 21.10% | 1-[(2R)-2-(4-fluorophenyl)morpholin-4-yl]ethan-1-one | 1-[(2R)-2-(4-fluorophenyl)morpholin-4-yl]ethan-1-one | XDS | 99.80% | 3.8 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 47.4 | - | No | 32.9 | 78.5 | 58.6 | - | |||||||
| 5res | 1.65 Å | 2020-03-25 | X-ray | T3V | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.14% | 22.63% | 11.90% | 1-{4-[(2-fluorophenyl)sulfonyl]piperazin-1-yl}ethan-1-one | 1-{4-[(2-fluorophenyl)sulfonyl]piperazin-1-yl}ethan-1-one | XDS | 98.30% | 6.0 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 58.5 | - | No | 58.8 | 64.3 | 68.5 | - | |||||||
| 5ret | 1.68 Å | 2020-03-25 | X-ray | T47 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.20% | 22.20% | 12.50% | 1-{4-[(3-chlorophenyl)methyl]piperazin-1-yl}ethan-1-one | 1-{4-[(3-chlorophenyl)methyl]piperazin-1-yl}ethan-1-one | XDS | 99.80% | 5.5 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 44.5 | - | No | 63.1 | 48.6 | 52.8 | - | |||||||
| 5reu | 1.69 Å | 2020-03-25 | X-ray | T4D | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.30% | 23.23% | 16.30% | 2-[(4-acetylpiperazin-1-yl)sulfonyl]benzonitrile | 2-[(4-acetylpiperazin-1-yl)sulfonyl]benzonitrile | XDS | 99.80% | 4.2 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 52.7 | - | No | 52.7 | 64.3 | 63.3 | - | |||||||
| 5rev | 1.60 Å | 2020-03-25 | X-ray | T4J | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.63% | 22.17% | 11.20% | N-[3-(thiomorpholine-4-carbonyl)phenyl]acetamide | N-[3-(thiomorpholine-4-carbonyl)phenyl]acetamide | XDS | 97.90% | 6.2 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 52.8 | - | No | 63.5 | 52.1 | 64.8 | - | |||||||
| 5rew | 1.55 Å | 2020-03-25 | X-ray | T4M | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.48% | 22.36% | 11.60% | N-[(1R)-1-(naphthalen-1-yl)ethyl]acetamide | N-[(1R)-1-(naphthalen-1-yl)ethyl]acetamide | XDS | 99.50% | 5.6 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 50.7 | - | No | 61.6 | 52.1 | 62.7 | - | |||||||
| 5rex | 2.07 Å | 2020-03-25 | X-ray | T4V | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.49% | 25.07% | 13.70% | 1-{4-[(naphthalen-1-yl)methyl]piperazin-1-yl}ethan-1-one | 1-{4-[(naphthalen-1-yl)methyl]piperazin-1-yl}ethan-1-one | XDS | 99.80% | 5.5 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 40.1 | - | No | 34.6 | 64.3 | 57.1 | - | |||||||
| 5rey | 1.96 Å | 2020-03-25 | X-ray | T4Y | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.58% | 27.35% | 20.90% | 1-{4-[(2-methylphenyl)methyl]-1,4-diazepan-1-yl}ethan-1-one | 1-{4-[(2-methylphenyl)methyl]-1,4-diazepan-1-yl}ethan-1-one | XDS | 99.60% | 3.6 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 33.1 | - | No | 17.5 | 55.9 | 68.8 | - | |||||||
| 5rez | 1.79 Å | 2020-03-25 | X-ray | T54 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.15% | 27.21% | 15.00% | (1R,2S)-2-(thiophen-3-yl)cyclopentane-1-carboxamide | (1R,2S)-2-(thiophen-3-yl)cyclopentane-1-carboxamide | XDS | 99.30% | 4.9 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 25.3 | - | No | 18.3 | 59.9 | 47.9 | - | |||||||
| 5rf0 | 1.65 Å | 2020-03-25 | X-ray | T5D | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.11% | 22.64% | 13.10% | [1-(pyridin-2-yl)cyclopentyl]methanol | [1-(pyridin-2-yl)cyclopentyl]methanol | XDS | 99.80% | 5.5 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 57.7 | - | No | 58.7 | 55.9 | 75.3 | - | |||||||
| 5rf1 | 1.73 Å | 2020-03-25 | X-ray | T5G | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.62% | 23.61% | 13.00% | 4-bromobenzene-1-sulfonamide | 4-bromobenzene-1-sulfonamide | XDS | 99.50% | 5.7 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 50.1 | - | No | 48.7 | 64.3 | 62.3 | - | |||||||
| 5rf2 | 1.53 Å | 2020-03-25 | X-ray | HVB | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.17% | 22.16% | 9.40% | 1-azanylpropylideneazanium | 1-azanylpropylideneazanium | XDS | 99.00% | 6.1 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 62.3 | - | No | 63.6 | 59.9 | 75.7 | - | |||||||
| 5rf3 | 1.50 Å | 2020-03-25 | X-ray | T5V | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.53% | 22.10% | 9.80% | pyrimidin-5-amine | pyrimidin-5-amine | XDS | 98.60% | 6.2 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 56.9 | - | No | 64.1 | 48.6 | 75.6 | - | |||||||
| 5rf4 | 1.61 Å | 2020-03-25 | X-ray | T5Y | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.00% | 22.49% | 10.70% | pyridin-2-ol | pyridin-2-ol | XDS | 99.20% | 5.9 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 55.2 | - | No | 60.5 | 52.1 | 72.5 | - | |||||||
| 5rf5 | 1.74 Å | 2020-03-25 | X-ray | HV2 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.76% | 23.06% | 14.60% | 1,1-bis(oxidanylidene)thietan-3-ol | 1,1-bis(oxidanylidene)thietan-3-ol | XDS | 99.80% | 5.4 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 48.2 | - | No | 54.5 | 55.9 | 61.2 | - | |||||||
| 5rf6 | 1.45 Å | 2020-03-25 | X-ray | NTG | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.06% | 21.74% | 9.30% | 5-(1,4-oxazepan-4-yl)pyridine-2-carbonitrile | 5-(1,4-oxazepan-4-yl)pyridine-2-carbonitrile | XDS | 98.10% | 5.4 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 50.8 | - | No | 67.6 | 48.6 | 60.3 | - | |||||||
| 5rf7 | 1.54 Å | 2020-03-25 | X-ray | T67 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.04% | 21.69% | 8.80% | 1-(4-methylpiperazin-1-yl)-2-(1H-pyrrolo[2,3-b]pyridin-3-yl)ethan-1-one | 1-(4-methylpiperazin-1-yl)-2-(1H-pyrrolo[2,3-b]pyridin-3-yl)ethan-1-one | XDS | 99.30% | 6.5 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 59.4 | - | No | 68.2 | 55.9 | 69.3 | - | |||||||
| 5rf8 | 1.44 Å | 2020-03-25 | X-ray | SFY | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.73% | 21.32% | 7.80% | 4-amino-N-(pyridin-2-yl)benzenesulfonamide | 4-amino-N-(pyridin-2-yl)benzenesulfonamide | XDS | 98.00% | 6.7 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 65.0 | - | No | 71.3 | 55.9 | 77.6 | - | |||||||
| 5rf9 | 1.43 Å | 2020-03-25 | X-ray | S7D | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.20% | 21.54% | 7.70% | 1-[(2~{S})-2-methylmorpholin-4-yl]-2-pyrazol-1-yl-ethanone | 1-[(2~{S})-2-methylmorpholin-4-yl]-2-pyrazol-1-yl-ethanone | XDS | 95.40% | 7.3 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 51.4 | - | No | 69.4 | 39.8 | 68.7 | - | |||||||
| 5rfa | 1.52 Å | 2020-03-25 | X-ray | JGY | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.67% | 21.51% | 7.80% | 1-methyl-N-{[(2S)-oxolan-2-yl]methyl}-1H-pyrazole-3-carboxamide | 1-methyl-N-{[(2S)-oxolan-2-yl]methyl}-1H-pyrazole-3-carboxamide | XDS | 98.80% | 7.1 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 59.0 | - | No | 69.6 | 51.8 | 71.1 | - | |||||||
| 5rfb | 1.48 Å | 2020-03-25 | X-ray | K3S | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.33% | 22.35% | 7.70% | N-[(1-methyl-1H-1,2,3-triazol-4-yl)methyl]ethanamine | N-[(1-methyl-1H-1,2,3-triazol-4-yl)methyl]ethanamine | XDS | 97.90% | 6.6 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 55.8 | - | No | 61.7 | 64.3 | 60.3 | - | |||||||
| 5rfc | 1.40 Å | 2020-03-25 | X-ray | K1Y | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.95% | 21.28% | 6.90% | methyl (2-methyl-4-phenyl-1,3-thiazol-5-yl)carbamate | methyl (2-methyl-4-phenyl-1,3-thiazol-5-yl)carbamate | XDS | 92.70% | 7.7 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 57.0 | - | No | 71.8 | 48.6 | 68.2 | - | |||||||
| 5rfd | 1.41 Å | 2020-03-25 | X-ray | T6J | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.76% | 21.10% | 7.90% | 2-[(methylsulfonyl)methyl]-1H-benzimidazole | 2-[(methylsulfonyl)methyl]-1H-benzimidazole | XDS | 94.20% | 6.6 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 69.5 | - | No | 73.2 | 64.3 | 77.0 | - | |||||||
| 5rfe | 1.46 Å | 2020-03-25 | X-ray | JGG | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.69% | 21.36% | 6.00% | N-[(4-cyanophenyl)methyl]morpholine-4-carboxamide | N-[(4-cyanophenyl)methyl]morpholine-4-carboxamide | XDS | 94.40% | 9.6 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 57.3 | - | No | 71.0 | 42.5 | 75.6 | - | |||||||
| 5rff | 1.78 Å | 2020-03-25 | X-ray | T6M | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.20% | 24.74% | 18.50% | 1-{4-[(4-chlorophenyl)sulfonyl]piperazin-1-yl}ethan-1-one | 1-{4-[(4-chlorophenyl)sulfonyl]piperazin-1-yl}ethan-1-one | XDS | 99.80% | 4.5 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 26.0 | - | No | 37.7 | 26.4 | 63.4 | - | |||||||
| 5rfg | 2.32 Å | 2020-03-25 | X-ray | T6V | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.21% | 30.65% | 36.10% | N-[(3S)-1,1-dioxo-2,3-dihydro-1H-1lambda~6~-thiophen-3-yl]-N-phenylacetamide | N-[(3S)-1,1-dioxo-2,3-dihydro-1H-1lambda~6~-thiophen-3-yl]-N-phenylacetamide | XDS | 99.80% | 2.6 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 22.5 | - | No | 6.3 | 42.5 | 71.1 | - | |||||||
| 5rfh | 1.58 Å | 2020-03-25 | X-ray | T6Y | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.86% | 23.68% | 10.60% | 1-{4-[(5-chlorothiophen-2-yl)methyl]piperazin-1-yl}ethan-1-one | 1-{4-[(5-chlorothiophen-2-yl)methyl]piperazin-1-yl}ethan-1-one | XDS | 98.40% | 5.1 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 25.7 | - | No | 48.2 | 22.9 | 55.8 | - | |||||||
| 5rfi | 1.69 Å | 2020-03-25 | X-ray | T71 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.33% | 24.23% | 11.60% | 1-{4-[(2,5-dimethylphenyl)sulfonyl]piperazin-1-yl}ethan-1-one | 1-{4-[(2,5-dimethylphenyl)sulfonyl]piperazin-1-yl}ethan-1-one | XDS | 98.50% | 5.3 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 37.8 | - | No | 42.7 | 45.4 | 63.6 | - | |||||||
| 5rfj | 1.80 Å | 2020-03-25 | X-ray | T7A | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.64% | 24.14% | 19.40% | N-(4-methoxy-1,3-benzothiazol-2-yl)acetamide | N-(4-methoxy-1,3-benzothiazol-2-yl)acetamide | XDS | 99.80% | 4.0 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 48.5 | - | No | 43.5 | 73.6 | 55.0 | - | |||||||
| 5rfk | 1.75 Å | 2020-03-25 | X-ray | T7D | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.75% | 23.48% | 18.10% | N-(1-acetylpiperidin-4-yl)benzamide | N-(1-acetylpiperidin-4-yl)benzamide | XDS | 99.50% | 4.2 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 47.6 | - | No | 50.3 | 59.9 | 60.3 | - | |||||||
| 5rfl | 1.64 Å | 2020-03-25 | X-ray | T7G | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.43% | 22.47% | 13.40% | 1-acetyl-N-(2-hydroxyphenyl)piperidine-4-carboxamide | 1-acetyl-N-(2-hydroxyphenyl)piperidine-4-carboxamide | XDS | 99.40% | 4.8 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 49.0 | - | No | 60.6 | 45.4 | 67.2 | - | |||||||
| 5rfm | 2.06 Å | 2020-03-25 | X-ray | T7J | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.79% | 25.72% | 21.60% | N-[(3R)-1,1-dioxo-2,3-dihydro-1H-1lambda~6~-thiophen-3-yl]-N-(4-methylphenyl)acetamide | N-[(3R)-1,1-dioxo-2,3-dihydro-1H-1lambda~6~-thiophen-3-yl]-N-(4-methylphenyl)acetamide | XDS | 99.80% | 4.5 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 37.9 | - | No | 28.9 | 68.7 | 54.2 | - | |||||||
| 5rfn | 1.80 Å | 2020-03-25 | X-ray | T7P | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.61% | 23.90% | 17.20% | N-[(3R)-1,1-dioxo-2,3-dihydro-1H-1lambda~6~-thiophen-3-yl]-N-(4-fluorophenyl)acetamide | N-[(3R)-1,1-dioxo-2,3-dihydro-1H-1lambda~6~-thiophen-3-yl]-N-(4-fluorophenyl)acetamide | XDS | 99.90% | 4.3 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 48.2 | - | No | 45.7 | 64.3 | 61.7 | - | |||||||
| 5rfo | 1.83 Å | 2020-03-25 | X-ray | T7S | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.49% | 27.75% | 23.50% | 1-[4-(piperidine-1-carbonyl)piperidin-1-yl]ethan-1-one | 1-[4-(piperidine-1-carbonyl)piperidin-1-yl]ethan-1-one | XDS | 99.90% | 3.4 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 11.4 | - | No | 15.4 | 37.3 | 40.2 | - | |||||||
| 5rfp | 2.03 Å | 2020-03-25 | X-ray | T7V | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.67% | 29.68% | 29.70% | N-[(1S)-1-(3-chlorophenyl)ethyl]acetamide | N-[(1S)-1-(3-chlorophenyl)ethyl]acetamide | XDS | 99.80% | 2.8 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 17.9 | - | No | 8.1 | 55.9 | 45.8 | - | |||||||
| 5rfq | 1.76 Å | 2020-03-25 | X-ray | T7Y | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.00% | 22.70% | 19.40% | N-[3-(2-oxopyrrolidin-1-yl)phenyl]acetamide | N-[3-(2-oxopyrrolidin-1-yl)phenyl]acetamide | XDS | 99.60% | 4.7 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 59.2 | - | No | 58.0 | 64.3 | 70.6 | - | |||||||
| 5rfr | 1.71 Å | 2020-03-25 | X-ray | T81 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.53% | 23.77% | 13.10% | 1-{4-[(5-bromothiophen-2-yl)methyl]piperazin-1-yl}ethan-1-one | 1-{4-[(5-bromothiophen-2-yl)methyl]piperazin-1-yl}ethan-1-one | XDS | 99.20% | 5.4 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 36.5 | - | No | 47.3 | 48.6 | 53.1 | - | |||||||
| 5rfs | 1.70 Å | 2020-03-25 | X-ray | T84 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.69% | 23.27% | 14.20% | 1-{4-[(thiophen-3-yl)methyl]piperazin-1-yl}ethan-1-one | 1-{4-[(thiophen-3-yl)methyl]piperazin-1-yl}ethan-1-one | XDS | 99.20% | 5.3 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 41.9 | - | No | 52.4 | 42.5 | 64.8 | - | |||||||
| 5rft | 1.58 Å | 2020-03-25 | X-ray | T8A | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.53% | 23.68% | 12.10% | 1-[(4S)-4-phenyl-3,4-dihydroisoquinolin-2(1H)-yl]ethan-1-one | 1-[(4S)-4-phenyl-3,4-dihydroisoquinolin-2(1H)-yl]ethan-1-one | XDS | 98.70% | 4.3 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 42.6 | - | No | 48.2 | 45.4 | 67.3 | - | |||||||
| 5rfu | 1.53 Å | 2020-03-25 | X-ray | T8D | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.53% | 20.98% | 9.40% | 1-{4-[(5-chlorothiophen-2-yl)sulfonyl]piperazin-1-yl}ethan-1-one | 1-{4-[(5-chlorothiophen-2-yl)sulfonyl]piperazin-1-yl}ethan-1-one | XDS | 99.20% | 6.8 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 57.9 | - | No | 74.3 | 52.1 | 63.9 | - | |||||||
| 5rfv | 1.48 Å | 2020-03-25 | X-ray | T8J | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.73% | 22.36% | 8.70% | 1-[4-(thiophene-2-carbonyl)piperazin-1-yl]ethan-1-one | 1-[4-(thiophene-2-carbonyl)piperazin-1-yl]ethan-1-one | XDS | 97.90% | 6.2 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 57.6 | - | No | 61.6 | 55.9 | 72.3 | - | |||||||
| 5rfw | 1.43 Å | 2020-03-25 | X-ray | T8M | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.95% | 22.32% | 5.80% | 1-{4-[(thiophen-2-yl)methyl]piperazin-1-yl}ethan-1-one | 1-{4-[(thiophen-2-yl)methyl]piperazin-1-yl}ethan-1-one | XDS | 97.90% | 9.0 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 31.7 | - | No | 61.9 | 16.6 | 60.8 | - | |||||||
| 5rfx | 1.55 Å | 2020-03-25 | X-ray | T8P | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.44% | 21.63% | 10.40% | 1-[4-(4-methoxyphenyl)piperazin-1-yl]ethan-1-one | 1-[4-(4-methoxyphenyl)piperazin-1-yl]ethan-1-one | XDS | 99.20% | 5.8 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 53.7 | - | No | 68.6 | 45.4 | 68.1 | - | |||||||
| 5rfy | 1.90 Å | 2020-03-25 | X-ray | T8S | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.98% | 27.67% | 24.00% | 1-acetyl-N-methyl-N-(propan-2-yl)piperidine-4-carboxamide | 1-acetyl-N-methyl-N-(propan-2-yl)piperidine-4-carboxamide | XDS | 99.30% | 3.6 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 34.1 | - | No | 15.8 | 68.7 | 59.7 | - | |||||||
| 5rfz | 1.68 Å | 2020-03-25 | X-ray | T8V | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.08% | 22.65% | 14.60% | N-(2-chloropyridin-3-yl)acetamide | N-(2-chloropyridin-3-yl)acetamide | XDS | 99.70% | 4.9 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 48.7 | - | No | 58.7 | 48.6 | 65.3 | - | |||||||
| 5rg0 | 1.72 Å | 2020-03-25 | X-ray | T8Y | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.81% | 23.59% | 12.80% | 1,1'-(piperazine-1,4-diyl)di(ethan-1-one) | 1,1'-(piperazine-1,4-diyl)di(ethan-1-one) | XDS | 99.10% | 5.3 | Diamond (I04-1) | REFMAC | 2020-03-15 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 47.7 | - | No | 49.1 | 64.3 | 57.2 | - | |||||||
| 6kl2 | 2.63 Å | 2020-03-25 | X-ray | - | MERS-CoV | Nucleocapsid | No functional ligands | 28.00% | 28.98% | 9.50% | - | HKL-2000 | 94.50% | 8.3 | NSRRC (BL13B1) | PHENIX | 2019-07-29 | 32105468 | - | - | - | - | N/A | - | - | Nucleoprotein - N-terminal domain | 22.9 | - | No | - | 10.0 | 84.9 | 26.0 | - | |||||||
| 6kl5 | 3.09 Å | 2020-03-25 | X-ray | DJO | MERS-CoV | Nucleocapsid | Functional ligand | 28.11% | 29.88% | 13.70% | (phenylmethyl) (2S)-2-(hydroxymethyl)-2,3-dihydroindole-1-carboxylate | (phenylmethyl) (2S)-2-(hydroxymethyl)-2,3-dihydroindole-1-carboxylate | HKL-2000 | 96.10% | 5.6 | NSRRC (BL13C1) | PHENIX | 2019-07-29 | 32105468 | - | - | - | - | N/A | - | - | Nucleoprotein - N-terminal domain | 25.5 | - | No | - | 7.5 | 91.6 | 27.2 | - | ||||||
| 6kl6 | 2.77 Å | 2020-03-25 | X-ray | DJU | MERS-CoV | Nucleocapsid | Functional ligand | 23.15% | 27.07% | 7.20% | N,N-dimethyl-1-(5-phenylmethoxy-1H-indol-3-yl)methanamine | N,N-dimethyl-1-(5-phenylmethoxy-1H-indol-3-yl)methanamine | HKL-2000 | 99.50% | 11.8 | SPring-8 (BL44XU) | PHENIX | 2019-07-29 | 32105468 | - | - | - | - | N/A | - | - | Nucleoprotein - N-terminal domain | - | - | No | - | 19.2 | - | 14.6 | - | ||||||
| 6w41 | 3.08 Å | 2020-03-25 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 22.26% | 24.33% | 13.60% | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 100.00% | 5.0 | APS (23-ID-D) | PHENIX | 2020-03-09 | 32245784 | - | - | http://covid19.bioreproducibility.org/static/data/6w41/6w41_refined.pdb | http://covid19.bioreproducibility.org/static/data/6w41/6w41_refined.mtz | 24.70% | Shifted the coordinates into the reference unit cell with ACHESYM. Flipped two peptide bonds (Tyr380 of the spike fragment and Ile98 of the antibody heavy chain). There is some evidence of the presence of another NAG group bound to Asn343 of the spike protein, but we did not attempt to model it. | The coordinates are NOT in a standardized location in the unit cell. Comparatively low-resolution structure that is highly idealized. | CR3022 Fab heavy chain; CR3022 Fab light chain; Spike protein S1 - receptor binding domain | 62.7 | minimal | Yes | Yes | - | 41.7 | 95.1 | 63.2 | - | ||||||
| 6w61 | 2.00 Å | 2020-03-25 | X-ray | SAM | SARS-CoV-2 | NSP10/NSP16 | Protein-protein complex | 17.06% | 19.30% | 13.90% | S-ADENOSYLMETHIONINE | S-ADENOSYLMETHIONINE | HKL-3000 | 100.00% | 37.1 | APS (19-ID) | PHENIX | 2020-03-15 | - | ZN | - | - | - | N/A | - | - | 2'-O-methyltransferase; Non-structural protein 10 | 59.8 | - | No | - | 86.1 | 39.8 | 68.1 | - | ||||||
| 6w63 | 2.10 Å | 2020-03-25 | X-ray | X77 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 14.99% | 22.14% | N/A | N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide | N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide | HKL-2000 | 99.30% | 17.2 | APS (21-ID-G) | PHENIX | 2020-03-16 | - | - | - | http://covid19.bioreproducibility.org/static/data/6w63/6w63_refined.pdb | http://covid19.bioreproducibility.org/static/data/6w63/6w63_refined.mtz | 25.01% | Placed the coordinates in a standardized location in the unit cell using ACHESYM. | The coordinates are NOT in a standardized location in the unit cell. | 3C-like proteinase | 67.3 | minimal | Yes | Yes | - | - | 63.8 | 93.3 | 52.8 | - | ||||
| 6w6y | 1.45 Å | 2020-03-25 | X-ray | AMP, MES | SARS-CoV-2 | NSP3: Macro | Functional ligand | 14.21% | 18.91% | 10.90% | ADENOSINE MONOPHOSPHATE; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | ADENOSINE MONOPHOSPHATE; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | HKL-3000 | 98.60% | 21.3 | APS (19-ID) | REFMAC | 2020-03-18 | 32939273 | - | - | - | - | N/A | - | - | Non-structural protein 3 - ADP ribose phosphatase (ADRP) domain (UNP residues 1024-1192) | 86.2 | - | No | - | 88.2 | 89.6 | 96.8 | - | ||||||
| 6w75 | 1.95 Å | 2020-03-25 | X-ray | NA, SAM, FMT | SARS-CoV-2 | NSP10/NSP16 | Protein-protein complex | 15.73% | 17.45% | 7.70% | S-ADENOSYLMETHIONINE; FORMIC ACID; SODIUM ION | SODIUM ION; S-ADENOSYLMETHIONINE; FORMIC ACID | HKL-3000 | 100.00% | 25.8 | APS (21-ID-F) | REFMAC | 2020-03-18 | 32994211 | NA; ZN | - | - | - | N/A | - | - | SARS-CoV-2 NSP16 - UNP residues 6799-7096; SARS-CoV-2 NSP10 - UNP residues 4254-4392 | 60.2 | - | No | 94.0 | 14.0 | 87.0 | - | |||||||
| 6yb7 | 1.25 Å | 2020-03-25 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 17.06% | 19.17% | 13.70% | - | DIALS | 97.10% | 9.7 | Diamond (I04-1) | BUSTER | 2020-03-16 | - | - | - | http://covid19.bioreproducibility.org/static/data/6yb7/6yb7_refined.pdb | http://covid19.bioreproducibility.org/static/data/6yb7/6yb7_refined.mtz | 16.30% | Re-refinement with individual anisotropic B factors. Fixed several rotamers. Added two water molecules. Substantial improvement of the refinement statistics and better electron density map; minimal changes to the model. | Refinement statistics and electron density map can be improved with minimal changes to the model. | Replicase polyprotein 1ab | 71.8 | minimal | Yes | Yes | - | - | 92.0 | 52.3 | 75.4 | - | |||||
| 5r8t | 1.27 Å | 2020-04-01 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.41% | 20.81% | 6.50% | - | XDS | 86.10% | 7.1 | Diamond (I04-1) | REFMAC | 2020-03-03 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 70.3 | - | No | - | - | 75.6 | 68.7 | 72.1 | - | ||||||
| 6m71 | 2.90 Å | 2020-04-01 | Cryo-EM | - | SARS-CoV-2 | NSP7/NSP8/NSP12 | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | PHENIX | 2020-03-16 | 32277040 | - | - | - | - | N/A | - | - | SARS-Cov-2 NSP 12; SARS-Cov-2 NSP 7; SARS-Cov-2 NSP 8 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6w9c | 2.70 Å | 2020-04-01 | X-ray | - | SARS-CoV-2 | NSP3: PLpro | No functional ligands | 23.50% | 27.91% | 14.00% | - | HKL-3000 | 57.30% | 4.9 | APS (19-ID) | REFMAC | 2020-03-22 | - | ZN | - | http://covid19.bioreproducibility.org/static/data/6w9c/6w9c_refined.pdb | http://covid19.bioreproducibility.org/static/data/6w9c/6w9c_refined.mtz | 29.80% | Re-refinement with added NCS restraints for chains A, B and C. Fixed a few rotamer outliers. Substantial improvement of the model was not possible due to the quality of electron density. | Very low completeness of the experimental data (57.3%). Poor quality of the electron density maps. Ambiguity of model building especially in the regions of C-terminal zinc-finger-like domains. | Papain-like proteinase | 27.5 | moderate | No | Yes | - | 14.4 | 66.2 | 49.9 | - | ||||||
| 6waq | 2.20 Å | 2020-04-01 | X-ray | NAG | SARS-CoV | Spike | Protein-protein complex | 20.26% | 23.59% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | MOSFLM | 99.80% | 6.2 | APS (19-ID) | PHENIX | 2020-03-25 | 32375025 | - | - | - | - | N/A | - | - | nanobody SARS VHH-72; Spike glycoprotein | 28.8 | - | No | - | 49.1 | 39.6 | 44.7 | - | |||||||
| 6l5t | 1.72 Å | 2020-04-08 | X-ray | - | SADS-CoV | NSP3: PLpro | No functional ligands | 17.93% | 21.16% | 11.60% | - | XDS | 100.00% | 16.7 | SSRF (BL19U1) | PHENIX | 2019-10-24 | 32216114 | ZN | - | - | - | N/A | - | - | Peptidase C16 | 68.0 | - | No | - | 72.7 | 46.4 | 92.2 | - | |||||||
| 6w9q | 2.05 Å | 2020-04-08 | X-ray | PO4 | SARS-CoV-2 | NSP9 | Functional ligand | 23.27% | 24.59% | N/A | PHOSPHATE ION | PHOSPHATE ION | XDS | 100.00% | 13.7 | Australian Synchrotron (MX2) | PHENIX | 2020-03-23 | 32592996 | - | - | - | - | N/A | - | - | 3C-like proteinase peptide, Non-structural protein 9 fusion | 26.0 | - | No | - | 39.4 | 37.7 | 50.4 | - | ||||||
| 6yi3 | - | 2020-04-08 | NMR | - | SARS-CoV-2 | Nucleocapsid | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2020-03-31 | 33264373 | - | - | - | - | N/A | - | - | Nucleoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7btf | 2.95 Å | 2020-04-08 | Cryo-EM | - | SARS-CoV-2 | NSP7/NSP8/NSP12 | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2020-04-01 | 32277040 | ZN | - | - | - | N/A | - | - | NSP12; NSP7; NSP8 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 5rg1 | 1.65 Å | 2020-04-15 | X-ray | T9J | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.19% | 21.52% | 14.30% | Nalpha-acetyl-N-(3-bromoprop-2-yn-1-yl)-L-tyrosinamide | Nalpha-acetyl-N-(3-bromoprop-2-yn-1-yl)-L-tyrosinamide | XDS | 99.70% | 5.1 | Diamond (I04-1) | REFMAC | 2020-03-18 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 59.5 | - | No | - | 69.6 | 64.3 | 59.7 | - | ||||||
| 5rg2 | 1.63 Å | 2020-04-15 | X-ray | T9M | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.31% | 22.83% | 12.70% | N~2~-acetyl-N-prop-2-en-1-yl-D-allothreoninamide | N~2~-acetyl-N-prop-2-en-1-yl-D-allothreoninamide | XDS | 99.70% | 5.9 | Diamond (I04-1) | REFMAC | 2020-03-18 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 47.3 | - | No | - | 56.7 | 55.9 | 57.2 | - | ||||||
| 5rg3 | 1.58 Å | 2020-04-15 | X-ray | T9P | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.51% | 21.50% | 9.00% | N~2~-acetyl-N~1~-prop-2-en-1-yl-L-aspartamide | N~2~-acetyl-N~1~-prop-2-en-1-yl-L-aspartamide | XDS | 99.60% | 6.9 | Diamond (I04-1) | REFMAC | 2020-03-18 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 49.0 | - | No | - | 69.7 | 52.1 | 51.4 | - | ||||||
| 5rgg | 2.26 Å | 2020-04-15 | X-ray | NZD | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.65% | 21.65% | 33.80% | 4-methyl-N-phenylpiperazine-1-carboxamide | 4-methyl-N-phenylpiperazine-1-carboxamide | XDS | 99.40% | 4.4 | Diamond (I04-1) | REFMAC | 2020-04-07 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 51.6 | - | No | - | 68.4 | 45.4 | 64.2 | - | ||||||
| 5rgh | 1.70 Å | 2020-04-15 | X-ray | U0M | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.54% | 20.76% | 11.10% | 5-fluoro-1-[(5-methyl-1,3,4-thiadiazol-2-yl)methyl]-1,2,3,6-tetrahydropyridine | 5-fluoro-1-[(5-methyl-1,3,4-thiadiazol-2-yl)methyl]-1,2,3,6-tetrahydropyridine | XDS | 99.60% | 8.4 | Diamond (I04-1) | REFMAC | 2020-04-07 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 59.0 | - | No | - | 76.1 | 52.1 | 64.3 | - | ||||||
| 5rgi | 1.57 Å | 2020-04-15 | X-ray | U0P | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.03% | 21.17% | 9.60% | N'-cyclopropyl-N-methyl-N-[(5-methyl-1,2-oxazol-3-yl)methyl]urea | N'-cyclopropyl-N-methyl-N-[(5-methyl-1,2-oxazol-3-yl)methyl]urea | XDS | 99.40% | 7.2 | Diamond (I04-1) | REFMAC | 2020-04-07 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 54.4 | - | No | - | 72.7 | 37.3 | 73.5 | - | ||||||
| 5rgj | 1.34 Å | 2020-04-15 | X-ray | U0S | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.57% | 20.62% | 7.10% | (5S)-7-(pyrazin-2-yl)-2-oxa-7-azaspiro[4.4]nonane | (5S)-7-(pyrazin-2-yl)-2-oxa-7-azaspiro[4.4]nonane | XDS | 87.80% | 8.0 | Diamond (I04-1) | REFMAC | 2020-04-07 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 53.1 | - | No | - | 77.1 | 45.4 | 58.6 | - | ||||||
| 5rgk | 1.43 Å | 2020-04-15 | X-ray | U0V | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.42% | 20.99% | 7.80% | 2-fluoro-N-[2-(pyridin-4-yl)ethyl]benzamide | 2-fluoro-N-[2-(pyridin-4-yl)ethyl]benzamide | XDS | 97.30% | 7.4 | Diamond (I04-1) | REFMAC | 2020-04-07 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 51.6 | - | No | - | 74.2 | 29.3 | 74.6 | - | ||||||
| 5rgl | 1.76 Å | 2020-04-15 | X-ray | U0Y | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.04% | 22.89% | 21.50% | 1-[4-(4-methylbenzene-1-carbonyl)piperazin-1-yl]ethan-1-one | 1-[4-(4-methylbenzene-1-carbonyl)piperazin-1-yl]ethan-1-one | XDS | 99.70% | 3.9 | Diamond (I04-1) | REFMAC | 2020-04-07 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 57.3 | - | No | - | 56.3 | 52.1 | 80.8 | - | ||||||
| 5rgm | 2.04 Å | 2020-04-15 | X-ray | U1D | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.39% | 22.37% | 16.40% | N'-acetyl-4,5,6,7-tetrahydro-1-benzothiophene-2-carbohydrazide | N'-acetyl-4,5,6,7-tetrahydro-1-benzothiophene-2-carbohydrazide | XDS | 99.90% | 6.1 | Diamond (I04-1) | REFMAC | 2020-04-07 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 48.0 | - | No | - | 61.6 | 59.9 | 49.7 | - | ||||||
| 5rgn | 1.86 Å | 2020-04-15 | X-ray | U1A | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.36% | 23.32% | 24.30% | 1-{4-[(4-methylphenyl)sulfonyl]piperazin-1-yl}ethan-1-one | 1-{4-[(4-methylphenyl)sulfonyl]piperazin-1-yl}ethan-1-one | XDS | 99.20% | 3.9 | Diamond (I04-1) | REFMAC | 2020-04-07 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 48.9 | - | No | - | 51.8 | 55.9 | 65.4 | - | ||||||
| 5rgo | 1.74 Å | 2020-04-15 | X-ray | U1G | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.49% | 21.75% | 15.50% | 1-[4-(furan-2-carbonyl)piperazin-1-yl]ethan-1-one | 1-[4-(furan-2-carbonyl)piperazin-1-yl]ethan-1-one | XDS | 99.60% | 5.3 | Diamond (I04-1) | REFMAC | 2020-04-07 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 49.7 | - | No | - | 67.5 | 37.3 | 69.7 | - | ||||||
| 5rgp | 2.07 Å | 2020-04-15 | X-ray | U1M | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.10% | 20.19% | 13.00% | 1-{4-[(2,4-dimethylphenyl)sulfonyl]piperazin-1-yl}ethan-1-one | 1-{4-[(2,4-dimethylphenyl)sulfonyl]piperazin-1-yl}ethan-1-one | XDS | 99.50% | 6.6 | Diamond (I04-1) | REFMAC | 2020-04-07 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 76.2 | - | No | - | 80.5 | 73.6 | 77.1 | - | ||||||
| 5rgq | 2.15 Å | 2020-04-15 | X-ray | U1V | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.00% | 22.50% | 11.00% | 1-(4-fluoro-2-methylphenyl)methanesulfonamide | 1-(4-fluoro-2-methylphenyl)methanesulfonamide | XDS | 99.80% | 8.4 | Diamond (I04-1) | REFMAC | 2020-04-07 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 48.4 | - | No | - | 60.1 | 35.0 | 77.0 | - | ||||||
| 5rgr | 1.41 Å | 2020-04-15 | X-ray | K1G | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.28% | 20.01% | 5.70% | N,1-dimethyl-N-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine | N,1-dimethyl-N-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine | XDS | 88.50% | 8.6 | Diamond (I04-1) | REFMAC | 2020-04-07 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 69.6 | - | No | - | 81.6 | 64.3 | 69.0 | - | ||||||
| 5rgs | 1.72 Å | 2020-04-15 | X-ray | S7V | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.25% | 23.06% | 13.40% | [(2~{R})-4-(phenylmethyl)morpholin-2-yl]methanol | [(2~{R})-4-(phenylmethyl)morpholin-2-yl]methanol | XDS | 99.50% | 6.2 | Diamond (I04-1) | REFMAC | 2020-04-07 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 50.5 | - | No | - | 54.5 | 55.9 | 65.7 | - | ||||||
| 6m2n | 2.20 Å | 2020-04-15 | X-ray | 3WL | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 22.54% | 25.44% | 9.20% | 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one | 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one | HKL-3000 | 97.80% | 4.9 | SSRF (BL18U1) | PHENIX | 2020-02-28 | 32737471 | - | - | http://covid19.bioreproducibility.org/static/data/6m2n/6m2n_refined.pdb | http://covid19.bioreproducibility.org/static/data/6m2n/6m2n_refined.mtz | 27.10% | Achesymed. Rotated residues: Gln 306A, His 41A, His 41B, His41C, His 164C, His 41D. Added few waters. | The coordinates are NOT in a standardized location in the unit cell. | SARS-CoV-2 3CL protease | 17.4 | minimal | Yes | Yes | - | 31.3 | 26.6 | 50.6 | - | |||||
| 6m2q | 1.70 Å | 2020-04-15 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 17.55% | 20.40% | 7.70% | - | HKL-2000 | 99.00% | 4.0 | SSRF (BL18U1) | PHENIX | 2020-02-28 | 32737471 | - | - | http://covid19.bioreproducibility.org/static/data/6m2q/6m2q_refined.pdb | http://covid19.bioreproducibility.org/static/data/6m2q/6m2q_refined.mtz | 21.80% | Placed the coordinates in a standardized location in the unit cell using ACHESYM. | The coordinates are NOT in a standardized location in the unit cell. | SARS-CoV-2 3CL protease | 81.6 | minimal | Yes | Yes | - | 78.8 | 73.6 | 96.1 | - | ||||||
| 6wcf | 1.06 Å | 2020-04-15 | X-ray | MES | SARS-CoV-2 | NSP3: Macro | No functional ligands | 12.54% | 15.37% | 9.40% | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | HKL-3000 | 97.90% | 18.74 | APS (19-ID) | PHENIX | 2020-03-30 | 32939273 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 67.1 | - | No | 97.9 | 38.1 | 73.3 | - | |||||||
| 6wen | 1.35 Å | 2020-04-15 | X-ray | - | SARS-CoV-2 | NSP3: Macro | No functional ligands | 10.36% | 14.41% | 8.90% | - | HKL-3000 | 95.70% | 24.13 | APS (19-ID) | REFMAC | 2020-04-02 | 32939273 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 87.4 | - | No | 98.8 | 95.7 | 99.6 | - | ||||||||
| 6yla | 2.42 Å | 2020-04-15 | X-ray | MLI, NAG, 1PE, PG0 | SARS-CoV-2 | Spike | Pathogen-host interaction | 21.27% | 23.67% | 30.30% | MALONATE ION; 2-(2-METHOXYETHOXY)ETHANOL; PENTAETHYLENE GLYCOL | MALONATE ION; 2-acetamido-2-deoxy-beta-D-glucopyranose; PENTAETHYLENE GLYCOL; 2-(2-METHOXYETHOXY)ETHANOL | xia2 | 100.00% | 11.6 | Diamond (I03) | PHENIX; PHENIX | 2020-04-06 | 32585135 | - | - | http://covid19.bioreproducibility.org/static/data/6yla/6yla_refined.pdb | http://covid19.bioreproducibility.org/static/data/6yla/6yla_refined.mtz | 23.80% | Replaced 18 buffer molecules with water molecules or PEG. | Multiple buffer molecules (DMS, MLE, MLI) with very weak electron density. | Spike glycoprotein; Heavy Chain; Light chain | 43.9 | minimal | Yes | - | 48.3 | 43.6 | 71.5 | - | ||||||
| 6wiq | 2.85 Å | 2020-04-22 | X-ray | - | SARS-CoV-2 | NSP7/NSP8 | Protein-protein complex | 20.82% | 25.23% | 13.00% | - | HKL-3000 | 97.90% | 18.2 | APS (19-ID) | PHENIX | 2020-04-10 | 34197805 | - | - | - | - | N/A | - | - | SARS-CoV-2 NSP7 - UNP residues 3860-3942; SARS-CoV-2 NSP8 - C-terminal domain (UNP residues 4019-4140) | 64.7 | - | No | - | 33.1 | 90.8 | 80.3 | - | |||||||
| 6wji | 2.05 Å | 2020-04-22 | X-ray | - | SARS-CoV-2 | Nucleocapsid | No functional ligands | 18.69% | 22.75% | 17.10% | - | HKL-3000 | 97.00% | 9.7 | APS (21-ID-F) | REFMAC | 2020-04-13 | 37191546 | - | - | - | - | N/A | - | - | SARS-CoV-2 nucleocapsid protein - C-terminal dimerization domain (UNP residues 257-364) | 67.1 | - | No | 57.6 | 82.1 | 69.5 | - | ||||||||
| 6wjt | 2.00 Å | 2020-04-22 | X-ray | NA, SAH, FMT | SARS-CoV-2 | NSP10/NSP16 | Functional ligand | 17.11% | 19.11% | 10.10% | SODIUM ION; FORMIC ACID; S-ADENOSYL-L-HOMOCYSTEINE | SODIUM ION; S-ADENOSYL-L-HOMOCYSTEINE; FORMIC ACID | HKL-3000 | 100.00% | 16.7 | APS (21-ID-F) | REFMAC | 2020-04-14 | 32994211 | ZN; NA | - | - | - | N/A | - | - | 2'-O-methyltransferase; Non-structural protein 10 | 59.3 | - | No | 87.1 | 14.9 | 91.1 | - | |||||||
| 7bqy | 1.70 Å | 2020-04-22 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.49% | 22.57% | 6.10% | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | - | HKL-2000 | 98.10% | 7.5 | SSRF (BL19U1) | PHENIX | 2020-03-26 | 32272481 | - | - | http://covid19.bioreproducibility.org/static/data/7bqy/7bqy_refined.pdb | http://covid19.bioreproducibility.org/static/data/7bqy/7bqy_refined.mtz | 23.70% | Deleted terminal benzyl group from the inhibitor (residue 010) and introduced full carboxyl COO including OXT atom in residue PJE. | Minor issues with inhibitor modeling. | main protease; N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | 49.2 | minimal | Yes | Yes | - | 59.6 | 35.0 | 78.9 | - | |||||
| 7bv1 | 2.80 Å | 2020-04-22 | Cryo-EM | - | SARS-CoV-2 | NSP7/NSP8/NSP12 | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | phenix.real_space_refine; PHENIX | 2020-04-09 | 32358203 | ZN | - | - | - | N/A | - | - | nsp12; nsp8; nsp7 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7bv2 | 2.50 Å | 2020-04-22 | Cryo-EM | POP, F86 | SARS-CoV-2 | NSP7/NSP8/NSP12 | Functional ligand | N/A | N/A | N/A | Templete; Primer; PYROPHOSPHATE 2-; [(2~{R},3~{S},4~{R},5~{R})-5-(4-azanylpyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-cyano-3,4-bis(oxidanyl)oxolan-2-yl]methyl dihydrogen phosphate | PYROPHOSPHATE 2-; [(2~{R},3~{S},4~{R},5~{R})-5-(4-azanylpyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-cyano-3,4-bis(oxidanyl)oxolan-2-yl]methyl dihydrogen phosphate | - | - | () | PHENIX; phenix.real_space_refine | 2020-04-09 | 32358203 | MG; ZN | - | - | - | N/A | - | - | nsp12; nsp8; nsp7; Primer; Templete | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 6l70 | 1.56 Å | 2020-04-29 | X-ray | K36 | PEDV | NSP5 (3CLpro) | Functional ligand | 17.41% | 20.70% | 5.50% | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | 98.60% | 17.0 | SSRF (BL17U) | PHENIX | 2019-10-30 | 32098094 | - | - | - | - | N/A | - | - | PEDV main protease | 57.0 | - | No | - | 76.5 | 33.4 | 78.7 | - | |||||||
| 6lze | 1.50 Å | 2020-04-29 | X-ray | FHR | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.60% | 19.89% | 4.20% | ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | XDS | 98.00% | 13.99 | NFPSS (BL19U1) | PHENIX | 2020-02-19 | 32321856 | - | - | http://covid19.bioreproducibility.org/static/data/6lze/6lze_refined.pdb | http://covid19.bioreproducibility.org/static/data/6lze/6lze_refined.mtz | 18.69% | Placed the coordinates in a standardized location in the unit cell using ACHESYM. Refined inhibitor. | The coordinates are NOT in a standardized location in the unit cell. Distortions in the inhibitor. | Replicase polyprotein 1ab | 55.9 | minimal | Yes | Yes | - | 82.5 | 23.9 | 80.1 | - | |||||
| 6m0k | 1.50 Å | 2020-04-29 | X-ray | FJC | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.95% | 19.31% | 3.00% | ~{N}-[(2~{S})-3-(3-fluorophenyl)-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | ~{N}-[(2~{S})-3-(3-fluorophenyl)-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | XDS | 98.80% | 18.69 | NFPSS (BL19U1) | PHENIX | 2020-02-22 | 32321856 | - | - | http://covid19.bioreproducibility.org/static/data/6m0k/6m0k_refined.pdb | http://covid19.bioreproducibility.org/static/data/6m0k/6m0k_refined.mtz | 17.80% | Placed the coordinates in a standardized location in the unit cell using ACHESYM. Refined inhibitor. | The coordinates are NOT in a standardized location in the unit cell. Distortions in the inhibitor. | Replicase polyprotein 1ab | 54.3 | minimal | Yes | Yes | - | 86.1 | 19.5 | 77.7 | - | |||||
| 6w37 | 2.90 Å | 2020-04-29 | X-ray | - | SARS-CoV-2 | ORF7a | No functional ligands | 23.59% | 26.80% | 24.90% | - | XDS | 99.90% | 9.8 | APS (21-ID-G) | PHENIX | 2020-03-09 | - | - | - | - | - | N/A | - | - | Protein 7a - H-2Kb residues 1-277 | 27.6 | - | No | - | - | 20.8 | 11.6 | 98.4 | - | ||||||
| 6wey | 0.95 Å | 2020-04-29 | X-ray | - | SARS-CoV-2 | NSP3: Macro | No functional ligands | 11.91% | 13.62% | 6.30% | - | DIALS | 98.20% | 13.1 | APS (21-ID-F) | PHENIX | 2020-04-03 | 32578982 | - | - | - | - | N/A | - | - | Non-structural protein 3 - Macro X domain (residues 207-377) | 83.9 | - | No | - | 99.3 | 63.8 | 96.0 | - | |||||||
| 6wkp | 2.67 Å | 2020-04-29 | X-ray | MES | SARS-CoV-2 | Nucleocapsid | No functional ligands | 19.73% | 24.77% | 9.10% | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | HKL-3000 | 97.50% | 8.2 | APS (19-ID) | PHENIX | 2020-04-16 | - | ZN | - | - | - | N/A | - | - | SARS-CoV-2 nucleocapsid protein - RNA-binding domain (UNP residues 47-173) | 55.0 | - | No | - | 37.5 | 68.6 | 78.5 | - | ||||||
| 6wkq | 1.98 Å | 2020-04-29 | X-ray | NA, SFG, FMT | SARS-CoV-2 | NSP10/NSP16 | Functional ligand | 16.23% | 17.99% | 7.10% | FORMIC ACID; SINEFUNGIN; SODIUM ION | SODIUM ION; SINEFUNGIN; FORMIC ACID | HKL-3000 | 100.00% | 22.2 | APS (21-ID-F) | REFMAC | 2020-04-16 | 32994211 | NA; ZN | - | - | - | N/A | - | - | SARS-CoV-2 NSP16 - UNP residues 6799-7096; SARS-CoV-2 NSP10 - UNP residues 4254-4392 | 63.1 | - | No | 92.2 | 13.7 | 94.9 | - | |||||||
| 6wlc | 1.82 Å | 2020-04-29 | X-ray | U5P, TRS, FMT | SARS-CoV-2 | NSP15 | Functional ligand | 17.03% | 19.49% | 14.70% | URIDINE-5'-MONOPHOSPHATE; 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL | URIDINE-5'-MONOPHOSPHATE; 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL; FORMIC ACID | HKL-3000 | 100.00% | 18.8 | APS (19-ID) | PHENIX | 2020-04-19 | 33564093 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 83.4 | - | No | - | 85.1 | 87.7 | 83.6 | - | ||||||
| 6yhu | 2.00 Å | 2020-04-29 | X-ray | - | SARS-CoV-2 | NSP7/NSP8 | Protein-protein complex | 21.43% | 23.93% | 7.41% | - | XDS | 99.39% | 17.1 | ROTATING ANODE () | PHENIX | 2020-03-31 | 32535228 | - | - | - | - | N/A | - | - | Replicase polyprotein 1a; Replicase polyprotein 1a | 69.9 | - | No | - | 45.5 | 70.3 | 99.7 | - | |||||||
| 6ym0 | 4.36 Å | 2020-04-29 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 32.28% | 31.88% | 68.30% | - | xia2 | 100.00% | 4.0 | Diamond (I03) | REFMAC | 2020-04-07 | 32585135 | - | - | - | - | N/A | - | - | Spike glycoprotein; heavy chain; light chain | 1.0 | - | No | - | 5.1 | 28.3 | 11.2 | - | |||||||
| 6ynq | 1.80 Å | 2020-04-29 | X-ray | P6N | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.22% | 22.61% | N/A | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one | XDS | 94.90% | 2.42 | PETRA III, DESY (P11) | PHENIX | 2020-04-14 | 33811162 | - | - | http://covid19.bioreproducibility.org/static/data/6ynq/6ynq_refined.pdb | http://covid19.bioreproducibility.org/static/data/6ynq/6ynq_refined.mtz | 24.70% | Linked the inhibitor to both conformations of Cys124. Placed the coordinates in a standardized location in the unit cell. Fixed conformations of the following residues: 1, 4, 5, 19, 22-25, 47, 55, 60, 61, 90, 117, 123, 128, 137, 154 (cut side-chain), 162, 165, 188, 205, 217, 221, 240, 254, 279, 284, 298, 304. Removed residues 305-306. Remodeled multiple water molecules. | The inhibitor at full occupancy is linked to only one of the two conformations of Cys145. Several poorly modelled side-chains. The coordinates are NOT in a standardized location in the unit cell. Discrepancy in reported resolution: 1.8 A author-reported on the main page, 1.3 A author-reported on the "experiment" page and the cif file, and the deposited data are at 1.75 A. Very skewed cumulative intensity distribution. | Replicase polyprotein 1ab | 39.2 | moderate | Yes | Yes | 59.0 | 48.9 | 46.4 | - | ||||||
| 6yor | 3.30 Å | 2020-04-29 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | PHENIX | 2020-04-15 | 32585135 | - | - | - | - | N/A | - | - | Spike glycoprotein; IgG H chain; IgG L chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7buy | 1.60 Å | 2020-04-29 | X-ray | JRY | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.93% | 20.12% | 3.40% | hexylcarbamic acid | hexylcarbamic acid | XDS | 99.20% | 17.25 | SSRF (BL17U1) | PHENIX | 2020-04-08 | 32382072 | - | - | http://covid19.bioreproducibility.org/static/data/7buy/7buy_refined.pdb | http://covid19.bioreproducibility.org/static/data/7buy/7buy_refined.mtz | 20.53% | Used ACHESYM to standardize location in the unit cell. | The coordinates are NOT in a standardized location in the unit cell. | SARS-CoV-2 virus Main protease | 41.7 | minimal | Yes | Yes | - | 80.9 | 26.6 | 51.9 | - | |||||
| 6wks | 1.80 Å | 2020-05-06 | X-ray | SAM, ADN, GTA | SARS-CoV-2 | NSP10/NSP16 | Protein-protein complex | 15.20% | 18.80% | 13.00% | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE; S-ADENOSYLMETHIONINE; ADENOSINE | S-ADENOSYLMETHIONINE; ADENOSINE; P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE | XDS | 99.90% | 13.6 | () | REFMAC | 2020-04-16 | 32709886 | ZN | - | - | - | N/A | - | - | 2'-O-methyltransferase; Non-structural protein 10 | 47.5 | - | No | - | 88.7 | 14.1 | 67.3 | - | ||||||
| 6wnp | 1.44 Å | 2020-05-06 | X-ray | U5G | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.07% | 19.59% | 6.43% | boceprevir (bound form) | boceprevir (bound form) | HKL-2000 | 99.00% | 19.85 | APS (21-ID-F) | PHENIX; PHENIX | 2020-04-23 | - | - | - | http://covid19.bioreproducibility.org/static/data/6wnp/6wnp_rerefinement.pdb | http://covid19.bioreproducibility.org/static/data/6wnp/6wnp_rerefinement.mtz | 15.84% | Used anisotropic refinement, modeled disorder for: LEU 67 A, GLN 110 A, SER 121 A, VAL 125 A, ASP 153 A, SER 267 A, MET 276 A. Chaged rotamers of: SER 1 A, ARG 76 A, GLN 127 A, THR 154 A, PRO 168 A, THR 196 A, LYS 236 A, ASN 277 A. Added 110 HOH molecules. | Missing disorder in few residues and a lot of unmodeled waters. | 3C-like proteinase | 57.1 | minimal | Yes | Yes | - | - | 84.5 | 28.0 | 76.4 | - | ||||
| 6woj | 2.20 Å | 2020-05-06 | X-ray | APR | SARS-CoV-2 | NSP3: Macro | Functional ligand | 19.93% | 25.19% | 13.00% | ADENOSINE-5-DIPHOSPHORIBOSE | ADENOSINE-5-DIPHOSPHORIBOSE | XDS | 99.40% | 7.3 | APS (17-ID) | PHENIX | 2020-04-24 | 33158944 | - | - | - | - | N/A | - | - | Non-structural protein 3 - UNP Residues 1023-1197 | 61.6 | - | No | - | 33.6 | 91.8 | 72.3 | - | ||||||
| 6wq3 | 2.10 Å | 2020-05-06 | X-ray | SAH, GTA, 8NK | SARS-CoV-2 | NSP10/NSP16 | Functional ligand | 16.61% | 18.59% | 7.50% | 7-methylguanosine 5'-diphosphate; S-ADENOSYL-L-HOMOCYSTEINE; P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE | S-ADENOSYL-L-HOMOCYSTEINE; P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE; 7-methylguanosine 5'-diphosphate | HKL-3000 | 98.80% | 21.1 | APS (21-ID-F) | REFMAC | 2020-04-28 | 32994211 | ZN | - | - | - | N/A | - | - | 2'-O-methyltransferase; Non-structural protein 10 | 61.5 | - | No | 89.8 | 12.8 | 94.9 | - | |||||||
| 6wqd | 1.95 Å | 2020-05-06 | X-ray | - | SARS-CoV-2 | NSP7/NSP8 | No functional ligands | 18.66% | 22.87% | 18.80% | - | HKL-3000 | 97.30% | 22.5 | APS (19-ID) | REFMAC; PHENIX | 2020-04-28 | 34197805 | - | - | - | - | N/A | - | - | SARS-CoV-2 NSP7 - UNP residues 3860-3942; SARS-CoV-2 NSP8 - C-terminal domain (UNP residues 4019-4140) | 62.8 | - | No | - | 56.4 | 63.4 | 80.4 | - | |||||||
| 6wqf | 2.30 Å | 2020-05-06 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 17.96% | 22.96% | 11.00% | - | CrysalisPro | 95.40% | 8.1 | ROTATING ANODE () | PHENIX | 2020-04-28 | 32581217 | - | - | http://covid19.bioreproducibility.org/static/data/6wqf/6wqf_refined_1.pdb | http://covid19.bioreproducibility.org/static/data/6wqf/6wqf_refined_1.mtz | 23.90% | Changed space group to C2, reindexed from h,k,l to -h-l, k, h. | Non-standard space group, while standard C2 would make it compatible with many other structures of this protein. | 3C-like proteinase | 61.9 | minimal | Yes | Yes | - | 55.5 | 45.7 | 97.3 | - | ||||||
| 6wrh | 1.60 Å | 2020-05-06 | X-ray | PO4 | SARS-CoV-2 | NSP3: PLpro | No functional ligands | 12.34% | 16.43% | 9.50% | PHOSPHATE ION | PHOSPHATE ION | HKL-3000 | 99.10% | 6.8 | APS (19-ID) | REFMAC | 2020-04-29 | 33531496 | ZN | - | - | - | N/A | - | - | Non-structural protein 3 | 77.3 | - | No | 96.3 | 44.0 | 94.0 | - | |||||||
| 6ywk | 2.20 Å | 2020-05-06 | X-ray | EPE | SARS-CoV-2 | NSP3: Macro | No functional ligands | 17.63% | 21.39% | 13.80% | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID | XDS | 100.00% | 10.7 | SLS (X06SA) | REFMAC | 2020-04-29 | 33850605 | MG | - | - | - | N/A | - | - | NSP3 macrodomain | 78.9 | - | No | - | 70.8 | 84.4 | 83.9 | - | ||||||
| 6ywl | 2.50 Å | 2020-05-06 | X-ray | APR | SARS-CoV-2 | NSP3: Macro | Functional ligand | 18.89% | 22.32% | 19.90% | ADENOSINE-5-DIPHOSPHORIBOSE | ADENOSINE-5-DIPHOSPHORIBOSE | XDS | 99.40% | 6.9 | SLS (X06SA) | REFMAC | 2020-04-29 | 33850605 | MG | - | - | - | N/A | - | - | NSP3 macrodomain | 72.5 | - | No | - | 61.9 | 79.2 | 80.6 | - | ||||||
| 6ywm | 2.16 Å | 2020-05-06 | X-ray | MES | SARS-CoV-2 | NSP3: Macro | No functional ligands | 17.52% | 22.92% | 16.20% | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | XDS | 100.00% | 8.3 | SLS (X06SA) | REFMAC | 2020-04-29 | 33850605 | MG | - | - | - | N/A | - | - | NSP3 macrodomain | 70.4 | - | No | - | 55.8 | 89.7 | 71.1 | - | ||||||
| 6wrz | 2.25 Å | 2020-05-13 | X-ray | SAH, GTA, MGP | SARS-CoV-2 | NSP10/NSP16 | Functional ligand | 16.20% | 19.02% | 7.70% | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE; P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE; S-ADENOSYL-L-HOMOCYSTEINE | S-ADENOSYL-L-HOMOCYSTEINE; P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE; 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE | HKL-3000 | 99.00% | 23.4 | APS (21-ID-F) | REFMAC | 2020-04-30 | 32994211 | ZN | - | - | - | N/A | - | - | 2'-O-methyltransferase; Non-structural protein 10 | 62.4 | - | No | 87.6 | 14.4 | 97.4 | - | |||||||
| 6wtc | 1.85 Å | 2020-05-13 | X-ray | ACY | SARS-CoV-2 | NSP7/NSP8 | No functional ligands | 18.68% | 21.41% | 8.30% | ACETIC ACID | HKL-3000 | 93.00% | 12.9 | APS (19-ID) | PHENIX | 2020-05-02 | - | - | - | - | - | N/A | - | - | SARS-CoV-2 NSP7 - UNP residues 3860-3942; SARS-CoV-2 NSP8 - C-terminal domain (UNP residues 4019-4140) | 78.2 | - | No | - | - | 70.5 | 82.4 | 84.0 | - | ||||||
| 6wvn | 2.00 Å | 2020-05-13 | X-ray | SAM, GTA, MGP, ADE | SARS-CoV-2 | NSP10/NSP16 | Functional ligand | 16.23% | 17.80% | 7.00% | ADENINE; S-ADENOSYLMETHIONINE; P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE; 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE | S-ADENOSYLMETHIONINE; P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE; 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE; ADENINE | HKL-3000 | 99.80% | 20.3 | APS (21-ID-F) | REFMAC | 2020-05-06 | 32994211 | ZN | - | - | - | N/A | - | - | 2'-O-methyltransferase; Non-structural protein 10 | 63.4 | - | No | 92.8 | 14.2 | 94.4 | - | |||||||
| 6yva | 3.18 Å | 2020-05-13 | X-ray | - | SARS-CoV-2 | NSP3: PLpro | Protein-protein complex | 24.96% | 29.02% | 5.75% | - | XDS | 99.61% | 8.58 | SLS (X06SA) | PHENIX; PHENIX | 2020-04-28 | 32726803 | ZN | - | - | - | N/A | - | - | Replicase polyprotein 1a; Ubiquitin-like protein ISG15 | 8.4 | - | No | - | 9.8 | 42.0 | 31.3 | - | |||||||
| 6yyt | 2.90 Å | 2020-05-13 | Cryo-EM | - | SARS-CoV-2 | NSP7/NSP8/NSP12/RNA | Functional ligand | N/A | N/A | N/A | RNA product | - | - | - | () | 2020-05-06 | 32438371 | ZN | - | - | - | N/A | - | - | nsp12; nsp8; nsp7; RNA product | N/A | - | No | - | N/A | N/A | N/A | RNA product | - | |||||||
| 6yz1 | 2.40 Å | 2020-05-13 | X-ray | SFG, MES | SARS-CoV-2 | NSP10/NSP16 | Functional ligand | 18.66% | 22.56% | 40.80% | SINEFUNGIN; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | SINEFUNGIN; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | XDS | 100.00% | 12.8 | LIQUID ANODE () | PHENIX | 2020-05-06 | 32709887 | ZN | - | - | - | N/A | - | - | nsp16; nsp10 | 62.8 | - | No | - | 59.7 | 87.6 | 53.1 | - | ||||||
| 7bro | 2.00 Å | 2020-05-13 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Protein-protein complex | 22.62% | 25.90% | N/A | - | HKL-2000 | 99.60% | 70.7 | SSRF (BL17U1) | PHENIX | 2020-03-29 | 32887884 | - | - | http://covid19.bioreproducibility.org/static/data/7bro/7bro-refmac.pdb | http://covid19.bioreproducibility.org/static/data/7bro/7bro-refmac.mtz | 24.50% | Re-refinement and rebuilding an incorrect model; move atoms into the electron density to reduce the amount of difference density. | In some places the main chain is twisted out of density. The coordinates are NOT in a standardized location in the unit cell. | 3C-like proteinase | 29.0 | moderate | Yes | Yes | - | 27.3 | 48.2 | 58.4 | - | ||||||
| 7brp | 1.80 Å | 2020-05-13 | X-ray | U5G | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 21.56% | 23.96% | N/A | boceprevir (bound form) | boceprevir (bound form) | HKL-2000 | 99.50% | 28.4 | SSRF (BL17U1) | PHENIX | 2020-03-29 | 32887884 | - | - | http://covid19.bioreproducibility.org/static/data/7brp/7brp_refine.pdb | http://covid19.bioreproducibility.org/static/data/7brp/7brp_refine.mtz | 21.90% | After re-refinement with standard TLS and minor corrections of model Rfree has dropped by 4%. | The coordinates are NOT in a standardized location in the unit cell. | 3C-like proteinase | 42.3 | minimal | Yes | Yes | - | 45.3 | 50.0 | 65.0 | - | |||||
| 7bz5 | 1.84 Å | 2020-05-13 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 16.71% | 19.14% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.41% | 16.9 | SSRF (BL17U1) | PHENIX | 2020-04-26 | 32404477 | - | - | - | - | N/A | - | - | Spike protein S1; Heavy chain of B38; Light chain of B38 | 64.3 | - | No | - | 87.0 | 29.5 | 86.8 | - | |||||||
| 6wtj | 1.90 Å | 2020-05-20 | X-ray | K36 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.86% | 23.48% | N/A | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | XDS | 97.90% | 7.18 | SSRL (BL12-2) | PHENIX | 2020-05-02 | 32855413 | - | - | http://covid19.bioreproducibility.org/static/data/6wtj/6wtj_rerefined.pdb | http://covid19.bioreproducibility.org/static/data/6wtj/6wtj_rerefined.mtz | 24.20% | Placed the coordinates in a standardized location in the unit cell. Modified residues 55, 151, 177, 222, 228, 235, 290, 298. Deleted 302-306. Changed Wat549 (special position) to Cl. Deleted Wat 523, 552. | Wrong number of reflections, the coordinates are not in a standard unit cell placement. | 3C-like proteinase | 27.9 | minimal | Yes | Yes | - | 50.3 | 42.8 | 38.5 | - | |||||
| 6wtk | 2.00 Å | 2020-05-20 | X-ray | UED | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.59% | 25.53% | N/A | N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | XDS | 96.00% | 5.6 | SSRL (BL12-2) | PHENIX | 2020-05-03 | 32855413 | - | - | http://covid19.bioreproducibility.org/static/data/6wtk/6wtk-refined.pdb | http://covid19.bioreproducibility.org/static/data/6wtk/6wtk_refined.mtz | 27.42% | Achesymed. The inhibitor was designated K36, as in other structures of GC376. | The data in the PDB apparently extend to 1.2 A, although only 2 A data were used for refinement. Higher resolution data are too weak to consider them to be observed. | 3C-like proteinase | - | minimal | Yes | Yes | - | 30.5 | - | 31.0 | - | |||||
| 6wtm | 1.85 Å | 2020-05-20 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 20.77% | 25.16% | N/A | - | XDS | 97.80% | 6.42 | SSRL (BL12-2) | PHENIX | 2020-05-03 | 32855413 | - | - | http://covid19.bioreproducibility.org/static/data/6wtm/6wtm_refined.pdb | http://covid19.bioreproducibility.org/static/data/6wtm/6wtm_refined.mtz | 25.70% | Placed the coordinates in a standardized location in the unit cell using ACHESYM. | The coordinates are NOT in a standardized location in the unit cell. | 3C-like proteinase | 53.3 | minimal | Yes | Yes | - | 33.8 | 83.7 | 63.9 | - | ||||||
| 6wtt | 2.15 Å | 2020-05-20 | X-ray | K36, B1S | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 21.45% | 30.01% | 11.00% | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | HKL-3000 | 100.00% | 5.2 | APS (19-ID) | REFMAC | 2020-05-03 | 32541865 | MG | - | http://covid19.bioreproducibility.org/static/data/6wtt/6wtt_refined.pdb | http://covid19.bioreproducibility.org/static/data/6wtt/6wtt_refined.mtz | 28.69% | Achesymed. | The coordinates are NOT in a standardized location in the unit cell. | 3C-like proteinase | 5.6 | minimal | Yes | Yes | - | 7.2 | 20.3 | 45.0 | - | |||||
| 6wuu | 2.79 Å | 2020-05-20 | X-ray | - | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 19.52% | 22.97% | 15.80% | VIR250 | - | XDS | 99.60% | 10.1 | APS (24-ID-C) | PHENIX | 2020-05-05 | 33067239 | MG; ZN | - | - | - | N/A | - | - | papain-like protease - UNP Residues 1563-1879; VIR250 | 64.2 | - | No | - | 55.4 | 77.2 | 70.7 | - | ||||||
| 6wx4 | 1.66 Å | 2020-05-20 | X-ray | - | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 16.96% | 19.58% | 9.40% | VIR251 | - | XDS | 99.50% | 15.5 | APS (24-ID-C) | PHENIX | 2020-05-09 | 33067239 | ZN | - | http://covid19.bioreproducibility.org/static/data/6wx4/6wx4_refined.pdb | http://covid19.bioreproducibility.org/static/data/6wx4/6wx4_refined.mtz | 18.10% | Corrected geometry of the inhibitor. Mutated Cys270 to Cso (hydroxy-cystein). Placed the coordinates in a standardized location in the unit cell. Altered conformations of the following residues: 2, 3,18, 60, 140, 270, 306, 308. Added 51 water molecules. | Incorrect geometry (flat instead of tetrahedral) of the inhibitor carbon atom linked to Cys111. Cys270 is oxidised and should be modelled as such. Many missing water molecules. Conformations of several residues can be corrected. The coordinates are NOT in a standardized location in the unit cell. | Non-structural protein 3; VIR251 | 85.6 | moderate | Yes | Yes | - | 84.5 | 89.2 | 95.5 | - | |||||
| 6wxc | 1.85 Å | 2020-05-20 | X-ray | CMU, PO4, FMT | SARS-CoV-2 | NSP15 | Functional ligand | 17.09% | 19.37% | 15.90% | FORMIC ACID; 5-CHLORO-6-(1-(2-IMINOPYRROLIDINYL) METHYL) URACIL; PHOSPHATE ION | 5-CHLORO-6-(1-(2-IMINOPYRROLIDINYL) METHYL) URACIL; PHOSPHATE ION; FORMIC ACID | HKL-3000 | 99.80% | 22.5 | APS (19-ID) | PHENIX | 2020-05-10 | 33564093 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 81.3 | - | No | - | 85.8 | 91.9 | 69.6 | - | ||||||
| 6wxd | 2.00 Å | 2020-05-20 | X-ray | - | SARS-CoV-2 | NSP9 | No functional ligands | 21.45% | 25.34% | N/A | - | XDS | 99.40% | 26.4 | Australian Synchrotron (MX2) | PHENIX | 2020-05-10 | 32592996 | - | - | - | - | N/A | - | - | Non-structural protein 9 | 11.9 | - | No | - | 32.2 | 18.2 | 43.8 | - | |||||||
| 6yun | 1.44 Å | 2020-05-20 | X-ray | - | SARS-CoV-2 | Nucleocapsid | No functional ligands | 14.79% | 18.88% | 3.40% | - | Aimless | 98.40% | 18.4 | SLS (X06DA) | PHENIX | 2020-04-27 | 33039147 | - | - | - | - | N/A | - | - | Nucleoprotein | 79.2 | - | No | - | 88.4 | 80.4 | 71.4 | - | |||||||
| 6yvf | 1.60 Å | 2020-05-20 | X-ray | A82, CA | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.69% | 20.81% | 7.30% | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid; CALCIUM ION | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid; CALCIUM ION | XDS | 95.20% | 7.48 | PETRA III, DESY (P11) | PHENIX | 2020-04-28 | 33811162 | CA | - | http://covid19.bioreproducibility.org/static/data/6yvf/6yvf_refined.pdb | http://covid19.bioreproducibility.org/static/data/6yvf/6yvf_refined.mtz | 24.29% | Placed the coordinates in a standardized location in the unit cell. | The coordinates are NOT in a standardized location in the unit cell. | Replicase polyprotein 1ab | 58.9 | minimal | Yes | Yes | - | 75.6 | 56.0 | 60.6 | - | |||||
| 7c22 | 2.00 Å | 2020-05-20 | X-ray | - | SARS-CoV-2 | Nucleocapsid | No functional ligands | 18.87% | 23.82% | 10.70% | - | XDS | 95.60% | 5.9 | SSRF (BL19U1) | REFMAC | 2020-05-07 | 35006385 | - | - | - | - | N/A | - | - | Nucleoprotein | 46.9 | - | No | - | 46.6 | 74.6 | 47.9 | - | |||||||
| 5rgt | 2.22 Å | 2020-05-27 | X-ray | UHS | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.61% | 27.14% | 19.60% | N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(5-tert-butyl-1,2-oxazol-3-yl)propanamide | N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(5-tert-butyl-1,2-oxazol-3-yl)propanamide | XDS | 99.80% | 5.2 | Diamond (I04-1) | REFMAC | 2020-05-15 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 57.2 | - | No | - | - | 18.7 | 78.5 | 91.7 | - | |||||
| 5rgu | 2.11 Å | 2020-05-27 | X-ray | UGD | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.81% | 23.83% | 13.90% | N-(3-{[(2R)-4-oxoazetidin-2-yl]oxy}phenyl)-2-(pyrimidin-5-yl)acetamide | N-(3-{[(2R)-4-oxoazetidin-2-yl]oxy}phenyl)-2-(pyrimidin-5-yl)acetamide | XDS | 99.70% | 5.8 | Diamond (I04-1) | BUSTER | 2020-05-15 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 65.4 | - | No | - | - | 46.5 | 88.6 | 70.6 | - | |||||
| 5rgv | 1.82 Å | 2020-05-27 | X-ray | UGG | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.71% | 23.19% | 6.20% | 2-(isoquinolin-4-yl)-N-phenylacetamide | 2-(isoquinolin-4-yl)-N-phenylacetamide | XDS | 99.70% | 7.6 | Diamond (I04-1) | BUSTER | 2020-05-15 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 60.4 | - | No | - | - | 53.3 | 73.6 | 68.5 | - | |||||
| 5rgw | 1.43 Å | 2020-05-27 | X-ray | UGM | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.99% | 20.33% | 4.50% | 2-(5-cyanopyridin-3-yl)-N-(pyridin-3-yl)acetamide | 2-(5-cyanopyridin-3-yl)-N-(pyridin-3-yl)acetamide | XDS | 98.00% | 8.4 | Diamond (I04-1) | BUSTER | 2020-05-15 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 76.4 | - | No | - | - | 79.4 | 64.3 | 88.1 | - | |||||
| 5rgx | 1.69 Å | 2020-05-27 | X-ray | UGP | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.24% | 21.81% | 8.70% | 2-(3-cyanophenyl)-N-(4-methylpyridin-3-yl)acetamide | 2-(3-cyanophenyl)-N-(4-methylpyridin-3-yl)acetamide | XDS | 98.20% | 6.0 | Diamond (I04-1) | BUSTER | 2020-05-15 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 64.4 | - | No | - | - | 66.9 | 52.1 | 84.7 | - | |||||
| 5rgy | 1.98 Å | 2020-05-27 | X-ray | UGS | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.47% | 24.61% | 22.20% | N-(4-methoxypyridin-2-yl)-2-(naphthalen-2-yl)acetamide | N-(4-methoxypyridin-2-yl)-2-(naphthalen-2-yl)acetamide | XDS | 99.40% | 3.4 | Diamond (I04-1) | BUSTER | 2020-05-15 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 44.2 | - | No | - | - | 38.9 | 68.7 | 56.2 | - | |||||
| 5rgz | 1.52 Å | 2020-05-27 | X-ray | UH1 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.01% | 20.90% | 4.90% | 2-(3-cyanophenyl)-N-(pyridin-3-yl)acetamide | 2-(3-cyanophenyl)-N-(pyridin-3-yl)acetamide | XDS | 96.90% | 8.6 | Diamond (I04-1) | BUSTER | 2020-05-15 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 69.0 | - | No | - | - | 74.8 | 68.7 | 70.0 | - | |||||
| 5rh0 | 1.92 Å | 2020-05-27 | X-ray | UH4 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.52% | 24.05% | 9.20% | N-(5-methylthiophen-2-yl)-N'-pyridin-3-ylurea | N-(5-methylthiophen-2-yl)-N'-pyridin-3-ylurea | XDS | 99.70% | 7.1 | Diamond (I04-1) | BUSTER | 2020-05-15 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 47.3 | - | No | - | - | 44.4 | 64.3 | 61.1 | - | |||||
| 5rh1 | 1.96 Å | 2020-05-27 | X-ray | UGV | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.40% | 24.29% | 15.60% | 2-(5-chlorothiophen-2-yl)-N-(pyridin-3-yl)acetamide | 2-(5-chlorothiophen-2-yl)-N-(pyridin-3-yl)acetamide | XDS | 99.70% | 4.2 | Diamond (I04-1) | BUSTER | 2020-05-15 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 60.3 | - | No | - | - | 42.2 | 83.5 | 69.4 | - | |||||
| 5rh2 | 1.83 Å | 2020-05-27 | X-ray | UH7 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.17% | 23.02% | 10.10% | 2-(3-chlorophenyl)-N-(4-methylpyridin-3-yl)acetamide | 2-(3-chlorophenyl)-N-(4-methylpyridin-3-yl)acetamide | XDS | 96.80% | 8.7 | Diamond (I04-1) | BUSTER | 2020-05-15 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 58.0 | - | No | - | - | 54.8 | 68.7 | 67.2 | - | |||||
| 5rh3 | 1.69 Å | 2020-05-27 | X-ray | UHA | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.76% | 23.17% | 9.10% | (2R)-2-(3-chlorophenyl)-N-(4-methylpyridin-3-yl)propanamide | (2R)-2-(3-chlorophenyl)-N-(4-methylpyridin-3-yl)propanamide | XDS | 99.30% | 6.5 | Diamond (I04-1) | BUSTER | 2020-05-15 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 62.0 | - | No | - | - | 53.4 | 68.7 | 76.3 | - | |||||
| 5rh4 | 1.34 Å | 2020-05-27 | X-ray | UHG | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.56% | 20.49% | 5.10% | (2R)-2-(6-chloro-9H-carbazol-2-yl)propanoic acid | (2R)-2-(6-chloro-9H-carbazol-2-yl)propanoic acid | XDS | 93.70% | 9.1 | Diamond (I04-1) | BUSTER | 2020-05-15 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 62.4 | - | No | - | - | 78.3 | 48.6 | 72.4 | - | |||||
| 5rh5 | 1.72 Å | 2020-05-27 | X-ray | UHV | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.60% | 22.29% | 8.90% | N-(5-tert-butyl-1,2-oxazol-3-yl)-N-[(1R)-2-[(4-methoxy-2-methylphenyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | N-(5-tert-butyl-1,2-oxazol-3-yl)-N-[(1R)-2-[(4-methoxy-2-methylphenyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | XDS | 99.80% | 8.3 | Diamond (I04-1) | BUSTER | 2020-05-15 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 64.4 | - | No | - | - | 62.4 | 64.3 | 76.9 | - | |||||
| 5rh6 | 1.60 Å | 2020-05-27 | X-ray | UHY | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.43% | 21.73% | 7.70% | N-[(1R)-2-[(2-ethyl-6-methylphenyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]-N-[6-(propan-2-yl)pyridin-3-yl]propanamide | N-[(1R)-2-[(2-ethyl-6-methylphenyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]-N-[6-(propan-2-yl)pyridin-3-yl]propanamide | XDS | 99.50% | 7.1 | Diamond (I04-1) | BUSTER | 2020-05-15 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 66.9 | - | No | - | - | 67.7 | 55.9 | 85.2 | - | |||||
| 5rh7 | 1.71 Å | 2020-05-27 | X-ray | UJ1 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.50% | 22.02% | 10.30% | N-(5-tert-butyl-1H-pyrazol-3-yl)-N-[(1R)-2-[(2-ethyl-6-methylphenyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | N-(5-tert-butyl-1H-pyrazol-3-yl)-N-[(1R)-2-[(2-ethyl-6-methylphenyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | XDS | 98.90% | 6.5 | Diamond (I04-1) | BUSTER | 2020-05-15 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 68.4 | - | No | - | - | 64.8 | 64.3 | 83.1 | - | |||||
| 5rh8 | 1.81 Å | 2020-05-27 | X-ray | UHM | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.74% | 22.05% | 19.20% | 2-(cyanomethoxy)-N-[(1,2-thiazol-4-yl)methyl]benzamide | 2-(cyanomethoxy)-N-[(1,2-thiazol-4-yl)methyl]benzamide | XDS | 99.40% | 3.8 | Diamond (I04-1) | BUSTER | 2020-05-15 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 75.4 | - | No | - | - | 64.6 | 88.6 | 75.6 | - | |||||
| 5rh9 | 1.91 Å | 2020-05-27 | X-ray | UJ4 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.23% | 22.50% | 21.00% | N-{4-[(1S)-1-methoxyethyl]phenyl}-N-[(1R)-2-[(4-methoxy-2-methylphenyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | N-{4-[(1S)-1-methoxyethyl]phenyl}-N-[(1R)-2-[(4-methoxy-2-methylphenyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | XDS | 99.50% | 4.0 | Diamond (I04-1) | BUSTER | 2020-05-15 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 64.2 | - | No | - | - | 60.1 | 78.5 | 64.5 | - | |||||
| 5rha | 1.51 Å | 2020-05-27 | X-ray | T8M | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.35% | 20.95% | 9.30% | 1-{4-[(thiophen-2-yl)methyl]piperazin-1-yl}ethan-1-one | 1-{4-[(thiophen-2-yl)methyl]piperazin-1-yl}ethan-1-one | XDS | 95.20% | 6.2 | Diamond (I04-1) | BUSTER | 2020-05-15 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 68.7 | - | No | - | - | 74.5 | 64.3 | 73.9 | - | |||||
| 6lnq | 2.24 Å | 2020-05-27 | X-ray | EJF | SARS-CoV | NSP5 (3CLpro) | Functional ligand | 19.92% | 23.91% | 12.20% | N-[(2S)-3-methyl-1-[[(2S)-4-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]pentan-2-yl]amino]-1-oxidanylidene-butan-2-yl]-1H-indole-2-carboxamide | N-[(2S)-3-methyl-1-[[(2S)-4-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]pentan-2-yl]amino]-1-oxidanylidene-butan-2-yl]-1H-indole-2-carboxamide | HKL-2000 | 99.90% | 16.88 | ROTATING ANODE () | PHENIX | 2020-01-01 | 32391184 | - | - | - | - | N/A | - | - | Severe Acute Respiratory Syndrome Coronavirus 3c Like Protease | 69.6 | - | No | - | 45.7 | 78.7 | 90.5 | - | ||||||
| 6lny | 2.25 Å | 2020-05-27 | X-ray | EOC | SARS-CoV | NSP5 (3CLpro) | Functional ligand | 20.41% | 25.27% | 9.00% | (2~{S})-4-methyl-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepiperidin-3-yl]propan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pentanamide | (2~{S})-4-methyl-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepiperidin-3-yl]propan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pentanamide | HKL-2000 | 98.70% | 21.68 | ROTATING ANODE () | PHENIX | 2020-01-02 | 32391184 | - | - | - | - | N/A | - | - | Replicase polyprotein 1a | 46.1 | - | No | - | 32.9 | 64.4 | 70.3 | - | ||||||
| 6lo0 | 1.94 Å | 2020-05-27 | X-ray | EOF | SARS-CoV | NSP5 (3CLpro) | Functional ligand | 22.73% | 26.84% | 10.60% | (2~{S})-4-methyl-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pentanamide | (2~{S})-4-methyl-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pentanamide | HKL-2000 | 99.50% | 19.21 | ROTATING ANODE () | PHENIX | 2020-01-02 | 32391184 | - | - | - | - | N/A | - | - | Replicase polyprotein 1a | 50.6 | - | No | - | 20.6 | 68.9 | 86.8 | - | ||||||
| 6m15 | 2.38 Å | 2020-05-27 | Cryo-EM | NAG | RH BAT CoV HKU2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-02-24 | 32555182 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6m16 | 2.83 Å | 2020-05-27 | Cryo-EM | NAG | SADS-CoV | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-02-24 | 32555182 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6wps | 3.10 Å | 2020-05-27 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-04-27 | 32422645 | - | - | - | - | N/A | - | - | spike glycoprotein - signaling sequence + ectodomain (UNP residues 14-1211) + foldon trimerization domain + TEV cleavage site + His-tag,signaling sequence + ectodomain (UNP residues 14-1211) + foldon trimerization domain + TEV cleavage site + His-tag; S309 neutralizing antibody heavy chain - Fab; S309 neutralizing antibody light chain - Fab | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6wpt | 3.70 Å | 2020-05-27 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-04-27 | 32422645 | - | - | - | - | N/A | - | - | spike glycoprotein - signaling sequence + ectodomain (UNP residues 14-1211) + foldon trimerization domain + TEV cleavage site + His-tag,signaling sequence + ectodomain (UNP residues 14-1211) + foldon trimerization domain + TEV cleavage site + His-tag; S309 neutralizing antibody heavy chain - Fab; S309 neutralizing antibody light chain - Fab | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6wzo | 1.42 Å | 2020-05-27 | X-ray | - | SARS-CoV-2 | Nucleocapsid | No functional ligands | 15.73% | 17.32% | N/A | - | XDS | 91.10% | 16.8 | APS (24-ID-E) | PHENIX | 2020-05-14 | 32654247 | - | - | - | - | N/A | - | - | Nucleoprotein | 86.0 | - | No | - | 94.3 | 88.1 | 90.4 | - | |||||||
| 6wzq | 1.45 Å | 2020-05-27 | X-ray | - | SARS-CoV-2 | Nucleocapsid | No functional ligands | 16.73% | 18.02% | 5.80% | - | XDS | 98.80% | 16.0 | APS (24-ID-E) | PHENIX | 2020-05-14 | 32654247 | - | - | - | - | N/A | - | - | Nucleoprotein | 67.3 | - | No | - | 92.1 | 36.7 | 81.0 | - | |||||||
| 6wzu | 1.79 Å | 2020-05-27 | X-ray | PO4 | SARS-CoV-2 | NSP3: PLpro | No functional ligands | 16.00% | 17.41% | 13.70% | PHOSPHATE ION | PHOSPHATE ION | HKL-3000 | 100.00% | 6.6 | APS (19-ID) | REFMAC | 2020-05-14 | 33531496 | ZN | - | - | - | N/A | - | - | Non-structural protein 3 | 66.5 | - | No | 94.1 | 32.3 | 81.7 | - | |||||||
| 6x1b | 1.97 Å | 2020-05-27 | X-ray | PO4 | SARS-CoV-2 | NSP15 | Functional ligand | 15.66% | 18.46% | 15.50% | DNA (5'-R(*GP*U)-3'); PHOSPHATE ION | PHOSPHATE ION | HKL-3000 | 100.00% | 16.0 | APS (19-ID) | PHENIX | 2020-05-18 | 33564093 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease; DNA (5'-R(*GP*U)-3') | 83.1 | - | No | - | 90.4 | 85.6 | 79.3 | - | ||||||
| 6x29 | 2.70 Å | 2020-05-27 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2020-05-20 | 32699321 | - | - | - | - | N/A | - | - | Spike glycoprotein - ectodomain (UNP residues 16-1208) | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6x2a | 3.30 Å | 2020-05-27 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2020-05-20 | 32699321 | - | - | - | - | N/A | - | - | Spike glycoprotein - ectodomain (UNP residues 16-1208) | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6x2b | 3.60 Å | 2020-05-27 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2020-05-20 | 32699321 | - | - | - | - | N/A | - | - | Spike glycoprotein - ectodomain (UNP residues 16-1208) | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6x2c | 3.20 Å | 2020-05-27 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2020-05-20 | 32699321 | - | - | - | - | N/A | - | - | Spike glycoprotein - ectodomain (UNP residues 16-1208) | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7bw4 | 3.70 Å | 2020-05-27 | Cryo-EM | - | SARS-CoV-2 | NSP7/NSP8/NSP12 | No functional ligands | N/A | N/A | N/A | - | - | - | () | PHENIX | 2020-04-13 | 32531208 | ZN | - | - | - | N/A | - | - | Replicase polyprotein 1ab; Replicase polyprotein 1ab; Replicase polyprotein 1ab | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7c01 | 2.88 Å | 2020-05-27 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 21.81% | 26.51% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.90% | 12.2 | SSRF (BL17U1) | PHENIX | 2020-04-29 | 32454512 | - | - | - | - | N/A | - | - | SARS-Cov-2 Spike protein S1; CB6 heavy chain; CB6 light chain | 9.9 | - | No | - | 22.8 | 31.3 | 33.8 | - | |||||||
| 7c2i | 2.50 Å | 2020-05-27 | X-ray | SAM | SARS-CoV-2 | NSP10/NSP16 | Functional ligand | 17.40% | 20.88% | 20.40% | S-ADENOSYLMETHIONINE | S-ADENOSYLMETHIONINE | HKL-2000 | 100.00% | 10.67 | SSRF (BL19U1) | PHENIX | 2020-05-07 | 32728018 | ZN | - | - | - | N/A | - | - | Replicase polyprotein 1ab - nsp16; Replicase polyprotein 1ab - nsp10 | 64.3 | - | No | - | 75.1 | 39.9 | 88.5 | - | ||||||
| 7c2j | 2.80 Å | 2020-05-27 | X-ray | SAM | SARS-CoV-2 | NSP10/NSP16 | Functional ligand | 20.21% | 23.49% | 29.50% | S-ADENOSYLMETHIONINE | S-ADENOSYLMETHIONINE | HKL-2000 | 99.90% | 6.67 | SSRF (BL19U1) | PHENIX | 2020-05-07 | 32728018 | ZN | - | - | - | N/A | - | - | Replicase polyprotein 1ab - nsp16; Replicase polyprotein 1ab - nsp10 | 50.7 | - | No | - | 50.2 | 46.2 | 80.0 | - | ||||||
| 6m1v | 1.50 Å | 2020-06-03 | X-ray | - | SARS-CoV-2 | Spike | No functional ligands | 20.34% | 21.20% | N/A | - | HKL-2000 | 100.00% | 49.0 | SSRF (BL17U1) | PHENIX | 2020-02-26 | 32482145 | - | - | - | - | N/A | - | - | spike protein | 72.4 | - | No | - | 72.3 | 52.0 | 97.1 | - | |||||||
| 6m3w | 3.90 Å | 2020-06-03 | Cryo-EM | NAG | SARS-CoV | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-03-04 | 32681106 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6x4i | 1.85 Å | 2020-06-03 | X-ray | U3P, NA | SARS-CoV-2 | NSP15 | Functional ligand | 16.58% | 18.89% | 14.60% | 3'-URIDINEMONOPHOSPHATE; SODIUM ION | 3'-URIDINEMONOPHOSPHATE; SODIUM ION | HKL-3000 | 100.00% | 6.3 | APS (19-ID) | PHENIX | 2020-05-22 | 33564093 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 82.3 | - | No | 88.4 | 72.3 | 90.8 | - | |||||||
| 6yz5 | 1.80 Å | 2020-06-03 | X-ray | NAG | SARS-CoV-2 | Spike | No functional ligands | 16.58% | 19.33% | 10.90% | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.00% | 20.6 | Diamond (I03) | REFMAC | 2020-05-06 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; Nanobody H11-D4 | 76.4 | - | No | - | - | 86.0 | 57.8 | 87.9 | - | ||||||
| 6yz7 | 3.30 Å | 2020-06-03 | X-ray | NAG | SARS-CoV-2 | Spike | No functional ligands | 23.77% | 26.88% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 98.00% | 4.9 | Diamond (I03) | REFMAC | 2020-05-06 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; Antibody Cr3022; Antibody light chain; Nanobody | 4.4 | - | No | - | - | 20.4 | 12.4 | 34.2 | - | ||||||
| 6z2m | 2.71 Å | 2020-06-03 | X-ray | NAG | SARS-CoV-2 | Spike | No functional ligands | 19.83% | 24.10% | 23.10% | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.00% | 7.4 | Diamond (I03) | PHENIX | 2020-05-17 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; CR3022 antibody; CR3022 antibody; nanobody D4 | 27.7 | - | No | - | - | 43.8 | 52.0 | 35.3 | - | ||||||
| 6z43 | 3.30 Å | 2020-06-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-05-22 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; Nanobody | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7bwj | 2.85 Å | 2020-06-03 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 21.91% | 26.40% | 15.40% | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.90% | 14.3 | SSRF (BL17U) | PHENIX | 2020-04-14 | 32454513 | - | - | - | - | N/A | - | - | SARS-CoV-2 receptor binding domain; antibody light chain; antibody heavy chain | 29.2 | - | No | - | 23.6 | 72.3 | 38.2 | - | |||||||
| 7bzf | 3.26 Å | 2020-06-03 | Cryo-EM | - | SARS-CoV-2 | NSP7/NSP8/NSP12/RNA | Functional ligand | N/A | N/A | N/A | RNA (31-MER); RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*CP*AP*CP*A)-3') | - | - | - | () | PHENIX | 2020-04-27 | 32526208 | ZN | - | - | - | N/A | - | - | RNA-directed RNA polymerase; Non-structural protein 8; Non-structural protein 7; RNA (31-MER); RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*CP*AP*CP*A)-3') | N/A | - | No | - | N/A | N/A | N/A | RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*CP*AP*CP*A)-3'); RNA (31-MER) | - | ||||||
| 7c2k | 2.93 Å | 2020-06-03 | Cryo-EM | - | SARS-CoV-2 | NSP7/NSP8/NSP12/RNA | Functional ligand | N/A | N/A | N/A | RNA (29-MER); RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*CP*AP*CP*AP*GP*G*(F86)P*G)-3') | - | - | - | () | PHENIX | 2020-05-07 | 32526208 | ZN | - | - | - | N/A | - | - | RNA-directed RNA polymerase; Non-structural protein 8; Non-structural protein 7; RNA (29-MER); RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*CP*AP*CP*AP*GP*G*(F86)P*G)-3') | N/A | - | No | - | N/A | N/A | N/A | RNA (29-MER); RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*CP*AP*CP*AP*GP*G*(F86)P*G)-3') | - | ||||||
| 5rhb | 1.43 Å | 2020-06-10 | X-ray | USD | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.58% | 20.69% | 12.70% | (E)-1-(pyrimidin-2-yl)methanimine | (E)-1-(pyrimidin-2-yl)methanimine | XDS | 97.10% | 5.2 | Diamond (I04-1) | BUSTER | 2020-05-16 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 69.8 | - | No | - | 76.7 | 55.9 | 82.6 | - | ||||||
| 5rhc | 1.58 Å | 2020-06-10 | X-ray | USA | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.11% | 21.73% | 9.80% | (E)-1-(1H-imidazol-2-yl)methanimine | (E)-1-(1H-imidazol-2-yl)methanimine | XDS | 97.10% | 6.2 | Diamond (I04-1) | BUSTER | 2020-05-16 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 64.4 | - | No | - | 67.7 | 55.9 | 80.0 | - | ||||||
| 5rhd | 1.57 Å | 2020-06-10 | X-ray | US7 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.22% | 20.59% | 8.60% | 1-[4-(methylsulfonyl)phenyl]piperazine | 1-[4-(methylsulfonyl)phenyl]piperazine | XDS | 99.50% | 7.1 | Diamond (I04-1) | BUSTER | 2020-05-16 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 72.8 | - | No | - | 77.5 | 73.6 | 71.3 | - | ||||||
| 5rhe | 1.56 Å | 2020-06-10 | X-ray | UPD | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.93% | 21.56% | 7.60% | 1-acetyl-N-(6-methoxypyridin-3-yl)piperidine-4-carboxamide | 1-acetyl-N-(6-methoxypyridin-3-yl)piperidine-4-carboxamide | XDS | 98.40% | 8.1 | Diamond (I04-1) | BUSTER | 2020-05-16 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 71.2 | - | No | - | 69.3 | 59.9 | 89.3 | - | ||||||
| 5rhf | 1.76 Å | 2020-06-10 | X-ray | UPJ | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.47% | 22.77% | 13.80% | 1-acetyl-N-methyl-N-phenylpiperidine-4-carboxamide | 1-acetyl-N-methyl-N-phenylpiperidine-4-carboxamide | XDS | 99.40% | 5.4 | Diamond (I04-1) | BUSTER | 2020-05-16 | 33028810 | - | - | - | - | N/A | - | - | 3C-like proteinase | 67.6 | - | No | - | 57.5 | 78.5 | 74.5 | - | ||||||
| 6x6p | 3.22 Å | 2020-06-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2020-05-28 | 32587972 | - | - | - | - | N/A | - | - | SARS-CoV-2 spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7byr | 3.84 Å | 2020-06-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-04-24 | 32425270 | - | - | - | - | N/A | - | - | SARS-CoV-2 Spike glycoprotein; Ab23-Fab-Heavy Chain; Ab23-Fab-Light Chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6m5i | 2.50 Å | 2020-06-17 | X-ray | - | SARS-CoV-2 | NSP7/NSP8 | No functional ligands | 20.80% | 29.17% | N/A | - | 99.70% | 8.01 | SSRF (BL17U) | PHENIX | 2020-03-10 | - | - | - | - | - | N/A | - | - | SARS-CoV-2 nsp8; SARS-CoV-2 nsp7 | 6.3 | - | No | - | - | 9.5 | 49.1 | 17.0 | - | |||||||
| 6xa4 | 1.65 Å | 2020-06-17 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.82% | 23.93% | 6.50% | inhibitor UAW241 | - | HKL-3000 | 98.20% | 8.2 | APS (19-ID) | REFMAC | 2020-06-03 | 33158912 | - | - | http://covid19.bioreproducibility.org/static/data/6xa4/6xa4_refined.pdb | http://covid19.bioreproducibility.org/static/data/6xa4/6xa4_refined.mtz | 24.80% | Achesymed, flipped His41 for proper H-bonding. | The coordinates are NOT in a standardized location in the unit cell. | main protease - UNP residues 3264-3569; inhibitor UAW241 | 56.2 | minimal | Yes | Yes | - | 45.5 | 60.2 | 81.4 | - | |||||
| 6xa9 | 2.90 Å | 2020-06-17 | X-ray | - | SARS-CoV-2 | NSP3: PLpro | No functional ligands | 19.97% | 23.05% | 16.30% | - | XDS | 100.00% | 10.5 | Australian Synchrotron (MX2) | PHENIX | 2020-06-04 | 32845033 | ZN | - | - | - | N/A | - | - | Papain-like protease - UNP residues 1563-1878; ISG15 CTD-propargylamide - C-terminal domain (UNP residues 79-157) | 40.5 | - | No | - | 54.5 | 22.9 | 79.3 | - | |||||||
| 6xaa | 2.70 Å | 2020-06-17 | X-ray | - | SARS-CoV-2 | NSP3: PLpro | No functional ligands | 20.96% | 25.99% | 15.20% | - | XDS | 98.10% | 6.8 | Australian Synchrotron (MX2) | PHENIX | 2020-06-04 | 32845033 | ZN | - | http://covid19.bioreproducibility.org/static/data/6xaa/6xaa_refined_tls.pdb | http://covid19.bioreproducibility.org/static/data/6xaa/6xaa_refined.mtz | 26.00% | Shifted the coordinates into the reference unit cell with ACHESYM. Added side-chains for residues: 23A, 25A, 30A, 43A, 47A, 53A, 60A, 65A, 70A, 143A, 218A, 24B. 291A - rotated. Added 18 water molecules | Many missing side-chains. Not in the standard position in the unit cell | Papain-like protease - UNP residues 1563-1878; Ubiquitin-propargylamide | 18.6 | moderate | Yes | Yes | - | 26.8 | 20.3 | 64.3 | - | ||||||
| 6xb0 | 1.80 Å | 2020-06-17 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 15.45% | 20.10% | N/A | - | CrysalisPro | 97.20% | 10.2 | ROTATING ANODE () | PHENIX | 2020-06-05 | 33063790 | - | - | http://covid19.bioreproducibility.org/static/data/6xb0/6xb0_refined.pdb | http://covid19.bioreproducibility.org/static/data/6xb0/6xb0_refined.mtz | 20.25% | Reindexed from I2 to C2 (-h-l,k,h); transformed by ACHESYM; changed DMSO to HOH. | Non-standard space group, while standard C2 would make it compatible with many other structures of this protein. | 3C-like proteinase | 71.5 | minimal | Yes | Yes | - | 81.0 | 52.2 | 86.1 | - | ||||||
| 6xb1 | 1.80 Å | 2020-06-17 | X-ray | NEN | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 15.87% | 20.19% | N/A | 1-ETHYL-PYRROLIDINE-2,5-DIONE | 1-ETHYL-PYRROLIDINE-2,5-DIONE | CrysalisPro | 94.80% | 10.0 | ROTATING ANODE () | PHENIX | 2020-06-05 | 33063790 | - | - | http://covid19.bioreproducibility.org/static/data/6xb1/6xb1_refined.pdb | http://covid19.bioreproducibility.org/static/data/6xb1/6xb1_refined.mtz | 21.02% | Reindexed from I2 to C2 (-h-l,k,h); transformed by ACHESYM; removed inhibitor NEN from Cys156. | Non-standard space group, while standard C2 would make it compatible with many other structures of this protein. | 3C-like proteinase | 72.6 | significant | Yes | Yes | - | 80.5 | 60.1 | 81.3 | - | |||||
| 6xb2 | 2.10 Å | 2020-06-17 | X-ray | NEN | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.56% | 25.72% | N/A | 1-ETHYL-PYRROLIDINE-2,5-DIONE | 1-ETHYL-PYRROLIDINE-2,5-DIONE | CrysalisPro | 99.90% | 11.3 | ROTATING ANODE () | PHENIX | 2020-06-05 | 33063790 | - | - | http://covid19.bioreproducibility.org/static/data/6xb2/6xb2_refined.pdb | http://covid19.bioreproducibility.org/static/data/6xb2/6xb2_refined.mtz | 28.23% | Reindexed from I2 to C2 (-h-l,k,h); transformed by ACHESYM; removed inhibitor NEN from Cys145 & Cys156; changed DMSO to HOH. | Non-standard space group, while standard C2 would make it compatible with many other structures of this protein. | 3C-like proteinase | 14.7 | significant | Yes | Yes | - | 28.9 | 26.6 | 46.5 | - | |||||
| 6xbg | 1.45 Å | 2020-06-17 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.14% | 20.57% | 8.60% | inhibitor UAW246; SODIUM ION | SODIUM ION | HKL-3000 | 98.70% | 9.5 | APS (19-ID) | REFMAC | 2020-06-05 | 33158912 | - | - | http://covid19.bioreproducibility.org/static/data/6xbg/6xbg_refined.pdb | http://covid19.bioreproducibility.org/static/data/6xbg/6xbg_refined.mtz | 18.10% | Achesymed, used anisotropic ADPs. | The coordinates are NOT in a standardized location in the unit cell. | main protease - UNP residues 3264-3569; inhibitor UAW246 | 47.3 | minimal | Yes | Yes | - | 77.6 | 24.4 | 67.8 | - | |||||
| 6xbh | 1.60 Å | 2020-06-17 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.35% | 22.07% | 6.80% | SODIUM ION; inhibitor UAW247 | SODIUM ION | HKL-3000 | 95.10% | 8.8 | APS (19-ID) | REFMAC | 2020-06-06 | 33158912 | - | - | http://covid19.bioreproducibility.org/static/data/6xbh/6xbh_refined.pdb | http://covid19.bioreproducibility.org/static/data/6xbh/6xbh_refined.mtz | 20.70% | Achesymed. Na changed to HOH. Single orientation of glycerol. His41 flipped. TLS applied (6 TLS groups). | The coordinates are NOT in a standardized location in the unit cell. | main protease - UNP residues 3264-3569; inhibitor UAW247 | 67.6 | minimal | Yes | Yes | - | 64.5 | 68.7 | 77.2 | - | |||||
| 6xbi | 1.70 Å | 2020-06-17 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.32% | 21.66% | 8.30% | inhibitor UAW248; SODIUM ION | SODIUM ION | HKL-3000 | 85.20% | 23.99 | APS (19-ID) | REFMAC | 2020-06-06 | 33158912 | - | - | http://covid19.bioreproducibility.org/static/data/6xbi/6xbi_refined.pdb | http://covid19.bioreproducibility.org/static/data/6xbi/6xbi_refined.mtz | 22.60% | Achesymed. | The coordinates are NOT in a standardized location in the unit cell. | main protease - UNP residues 3264-3569; inhibitor UAW248 | 73.8 | minimal | Yes | Yes | - | 68.4 | 83.7 | 72.8 | - | |||||
| 6xch | 2.20 Å | 2020-06-17 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.70% | 23.70% | N/A | Leupeptin | - | CrysalisPro | 98.30% | 6.3 | ROTATING ANODE () | PHENIX | 2020-06-08 | 33152262 | - | - | http://covid19.bioreproducibility.org/static/data/6xch/6xch_refined.pdb | http://covid19.bioreproducibility.org/static/data/6xch/6xch_refined.mtz | 24.03% | Reindexed from I2 to C2 (-h-l,k,h); transformed by ACHESYM. | Non-standard space group, while standard C2 would make it compatible with many other structures of this protein. | 3C-like proteinase; Leupeptin | 45.5 | minimal | Yes | Yes | - | 47.7 | 45.9 | 72.7 | - | |||||
| 6xdc | 2.90 Å | 2020-06-17 | Cryo-EM | - | SARS-CoV-2 | ORF3a | No functional ligands | N/A | N/A | N/A | - | - | - | () | PHENIX; phenix.real_space_refine | 2020-06-10 | 34158638 | - | - | - | - | N/A | - | - | Protein 3a | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6xdh | 2.35 Å | 2020-06-17 | X-ray | FMT | SARS-CoV-2 | NSP15 | No functional ligands | 15.72% | 18.25% | 7.70% | FORMIC ACID | FORMIC ACID | XDS | 100.00% | 22.57 | APS (21-ID-F) | PHENIX | 2020-06-10 | - | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease - BewuA.18928.a.MX151 | 80.0 | - | No | - | 91.2 | 62.3 | 89.3 | - | ||||||
| 6z2e | 1.70 Å | 2020-06-17 | X-ray | Q5T, NA | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.20% | 24.34% | 7.40% | (4~{S})-4-[[(2~{S})-2-[[(2~{S})-2-[[(2~{S})-2-[3-[2-[2-[2-[2-[5-[(3~{a}~{S},4~{R},6~{a}~{R})-2-oxidanylidene-3,3~{a},4,6~{a}-tetrahydro-1~{H}-thieno[3,4-d]imidazol-4-yl]pentanoylamino]ethoxy]ethoxy]ethoxy]ethoxy]propanoylamino]butanoyl]amino]-3,3-dimethyl-butanoyl]amino]-4-methyl-pentanoyl]amino]-6-methylsulfonyl-hexanamide; SODIUM ION | (4~{S})-4-[[(2~{S})-2-[[(2~{S})-2-[[(2~{S})-2-[3-[2-[2-[2-[2-[5-[(3~{a}~{S},4~{R},6~{a}~{R})-2-oxidanylidene-3,3~{a},4,6~{a}-tetrahydro-1~{H}-thieno[3,4-d]imidazol-4-yl]pentanoylamino]ethoxy]ethoxy]ethoxy]ethoxy]propanoylamino]butanoyl]amino]-3,3-dimethyl-butanoyl]amino]-4-methyl-pentanoyl]amino]-6-methylsulfonyl-hexanamide; SODIUM ION | XDS | 100.00% | 32.4 | PETRA III, DESY (P11) | REFMAC | 2020-05-15 | - | - | - | http://covid19.bioreproducibility.org/static/data/6z2e/6z2e_refined.pdb | http://covid19.bioreproducibility.org/static/data/6z2e/6z2e_refined.mtz | 25.50% | Achesymed. Na 404 removed. Cl 403 changed to HOH. Q5T from C42 removed. Inserted EDO at the end of the inhibitor. | The coordinates are NOT in a standardized location in the unit cell. | Main Protease | 49.1 | minimal | Yes | Yes | - | - | 41.6 | 56.1 | 75.6 | - | ||||
| 6z4u | 1.95 Å | 2020-06-17 | X-ray | 15P | SARS-CoV-2 | ORF9B | No functional ligands | 21.75% | 25.67% | 12.30% | POLYETHYLENE GLYCOL (N=34) | POLYETHYLENE GLYCOL (N=34) | DIALS | 99.30% | 8.5 | NSLS-II (17-ID-2) | REFMAC | 2020-05-25 | - | - | - | - | - | N/A | - | - | Protein 9b | 28.8 | - | No | - | - | 29.4 | 55.3 | 48.7 | - | |||||
| 7bq7 | 2.37 Å | 2020-06-17 | X-ray | SAM | SARS-CoV-2 | NSP10/NSP16 | Protein-protein complex | 17.98% | 20.85% | N/A | S-ADENOSYLMETHIONINE | S-ADENOSYLMETHIONINE | HKL-3000 | 99.70% | 18.8 | SSRF (BL17U) | PHENIX | 2020-03-24 | - | ZN | - | - | - | N/A | - | - | SARS-CoV-2 nsp16; SARS-CoV-2 nsp10 | 46.4 | - | No | - | - | 75.3 | 48.2 | 44.6 | - | |||||
| 7c8r | 2.30 Å | 2020-06-17 | X-ray | TG3 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.02% | 24.83% | N/A | ethyl (4R)-4-[[(2S)-4-methyl-2-[[(2S,3R)-3-[(2-methylpropan-2-yl)oxy]-2-(phenylmethoxycarbonylamino)butanoyl]amino]pentanoyl]amino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | ethyl (4R)-4-[[(2S)-4-methyl-2-[[(2S,3R)-3-[(2-methylpropan-2-yl)oxy]-2-(phenylmethoxycarbonylamino)butanoyl]amino]pentanoyl]amino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | HKL-3000 | 99.80% | 22.9 | NSRRC (TPS 05A) | REFMAC | 2020-06-03 | - | - | - | http://covid19.bioreproducibility.org/static/data/7c8r/7c8r_refined.pdb | http://covid19.bioreproducibility.org/static/data/7c8r/7c8r_refined.mtz | 26.10% | Very minimal adjustments to bonds and angles of the inhibitor only. | Very minimal adjustments to bonds and angles of the inhibitor required. | 3C-like proteinase | 17.3 | minimal | Yes | Yes | - | - | 36.8 | 37.0 | 34.5 | - | ||||
| 7c8t | 2.05 Å | 2020-06-17 | X-ray | NOL | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.43% | 23.34% | N/A | N-[(BENZYLOXY)CARBONYL]-O-(TERT-BUTYL)-L-THREONYL-3-CYCLOHEXYL-N-[(1S)-2-HYDROXY-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}ETHYL]-L-ALANINAMIDE | N-[(BENZYLOXY)CARBONYL]-O-(TERT-BUTYL)-L-THREONYL-3-CYCLOHEXYL-N-[(1S)-2-HYDROXY-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}ETHYL]-L-ALANINAMIDE | HKL-2000 | 99.90% | 39.5 | NSRRC (TPS 05A) | REFMAC | 2020-06-03 | - | - | - | http://covid19.bioreproducibility.org/static/data/7c8t/7c8t_refined.pdb | http://covid19.bioreproducibility.org/static/data/7c8t/7c8t_refined.mtz | 25.20% | Achesymed, changed TLS to 6 groups. | The coordinates are NOT in a standardized location in the unit cell. | 3C-like proteinase | 21.0 | minimal | Yes | Yes | - | - | 51.6 | 19.2 | 45.9 | - | ||||
| 6xdg | 3.90 Å | 2020-06-24 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | PHENIX | 2020-06-10 | 32540901 | - | - | - | - | N/A | - | - | Spike glycoprotein - receptor binding domain (UNP residues 319-541); REGN10933 antibody Fab fragment light chain; REGN10933 antibody Fab fragment heavy chain; REGN10987 antibody Fab fragment heavy chain; REGN10987 antibody Fab fragment light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6xfn | 1.70 Å | 2020-06-24 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.54% | 22.82% | 6.50% | UAW243 | - | HKL-3000 | 98.90% | 8.7 | APS (19-ID) | REFMAC | 2020-06-15 | 33158912 | - | - | http://covid19.bioreproducibility.org/static/data/6xfn/6xfn_refined.pdb | http://covid19.bioreproducibility.org/static/data/6xfn/6xfn_refined.mtz | 23.33% | Achesymed and added TLS. The incorrect chirality of the P1’ norleucine of the inhibitor was corrected. | Incorrect chirality of the P1’ norleucine of the inhibitor. The coordinates are NOT in a standardized location in the unit cell. | main protease - UNP residues 3264-3569; UAW243 | 53.4 | minimal | Yes | Yes | - | 56.8 | 56.0 | 68.9 | - | |||||
| 6xg3 | 2.48 Å | 2020-06-24 | X-ray | PO4 | SARS-CoV-2 | NSP3: PLpro | No functional ligands | 15.13% | 19.30% | 16.70% | PHOSPHATE ION | PHOSPHATE ION | HKL-3000 | 100.00% | 3.7 | APS (19-ID) | REFMAC | 2020-06-16 | 33531496 | ZN | - | - | - | N/A | - | - | Peptidase C16 | 69.6 | - | No | 86.1 | 46.6 | 82.0 | - | |||||||
| 6zct | 2.55 Å | 2020-06-24 | X-ray | - | SARS-CoV-2 | NSP10 | No functional ligands | 18.95% | 19.46% | 11.40% | - | XDS | 100.00% | 11.42 | MAX IV (BioMAX) | PHENIX | 2020-06-12 | 33036230 | ZN | - | - | - | N/A | - | - | nsp10 | 68.1 | - | No | - | 85.2 | 53.1 | 73.3 | - | |||||||
| 6zcz | 2.65 Å | 2020-06-24 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 21.46% | 25.95% | 20.90% | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 99.20% | 5.0 | Diamond (I03) | PHENIX; PHENIX | 2020-06-12 | 32737466 | MG | - | - | - | N/A | - | - | Spike glycoprotein; Nanobody; EY6A heavy chain; EY6A light chain | 23.7 | - | No | - | 27.0 | 30.8 | 64.8 | - | |||||||
| 6zer | 3.80 Å | 2020-06-24 | X-ray | NAG, PO4 | SARS-CoV-2 | Spike | Protein-protein complex | 21.19% | 25.11% | 22.70% | PHOSPHATE ION | 2-acetamido-2-deoxy-beta-D-glucopyranose; PHOSPHATE ION | xia2 | 100.00% | 7.3 | Diamond (I03) | PHENIX | 2020-06-16 | 32737466 | - | - | - | - | N/A | - | - | RBD; heavy chain; light chain | 13.0 | - | No | - | 34.1 | 14.7 | 48.4 | - | ||||||
| 7c8u | 2.35 Å | 2020-06-24 | X-ray | K36 | SARS-CoV-2 | NSP5 (3CLpro) | Protein-protein complex | 21.15% | 27.28% | 6.50% | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | XDS | 99.20% | 9.7 | CLSI (08ID-1) | REFMAC | 2020-06-03 | - | - | - | http://covid19.bioreproducibility.org/static/data/7c8u/7c8u_refined.pdb | http://covid19.bioreproducibility.org/static/data/7c8u/7c8u_refined.mtz | N/A | - | - | Replicase polyprotein 1ab | 27.2 | - | Yes | - | - | 18.0 | 42.1 | 69.8 | - | |||||
| 7c8v | 2.15 Å | 2020-06-24 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 18.36% | 22.39% | 16.30% | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 100.00% | 10.2 | SSRF (BL19U1) | PHENIX; PHENIX | 2020-06-03 | 34330908 | - | - | - | - | N/A | - | - | Synthetic nanobody SR4; Spike glycoprotein - Receptor binding domain (RBD) | 68.1 | - | No | - | 61.4 | 79.2 | 71.0 | - | |||||||
| 7c8w | 2.77 Å | 2020-06-24 | X-ray | - | SARS-CoV-2 | Spike | Protein-protein complex | 20.50% | 26.68% | 27.60% | - | XDS | 100.00% | 11.1 | SSRF (BL19U1) | PHENIX; PHENIX | 2020-06-03 | 34330908 | - | - | - | - | N/A | - | - | Synthetic nanobody MR17; Spike glycoprotein - Receptor binding domain | 16.9 | - | No | - | 21.8 | 41.1 | 44.4 | - | |||||||
| 7can | 2.94 Å | 2020-06-24 | X-ray | - | SARS-CoV-2 | Spike | Protein-protein complex | 21.44% | 26.71% | 22.70% | - | XDS | 99.90% | 12.4 | SSRF (BL19U1) | PHENIX | 2020-06-09 | 34330908 | - | - | - | - | N/A | - | - | sybody MR17-K99Y; Spike glycoprotein - Receptor-binding domain (RBD) | 14.1 | - | No | - | 21.4 | 40.7 | 38.3 | - | |||||||
| 6xcm | 3.42 Å | 2020-07-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-06-08 | 32645326 | - | - | - | - | N/A | - | - | Spike glycoprotein; C105 Fab Heavy Chain; C105 Fab Light Chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6xcn | 3.66 Å | 2020-07-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-06-08 | 32645326 | - | - | - | - | N/A | - | - | Spike glycoprotein; C105 Fab Heavy Chain; C105 Fab Light Chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6xe1 | 2.75 Å | 2020-07-01 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 21.16% | 23.94% | 3.07% | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.93% | 13.01 | APS (19-ID) | PHENIX | 2020-06-11 | 33110068 | - | - | - | - | N/A | - | - | CV30 Fab Heavy chain; CV30 Fab Kappa chain; Spike protein S1 - receptor binding domain | 36.5 | - | No | - | 45.4 | 33.5 | 70.1 | - | |||||||
| 6xhu | 1.80 Å | 2020-07-01 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 20.21% | 24.64% | N/A | - | CrysalisPro | 99.20% | 0.026 | ROTATING ANODE () | PHENIX | 2020-06-19 | 33063790 | - | - | http://covid19.bioreproducibility.org/static/data/6xhu/6xhu_refined.pdb | http://covid19.bioreproducibility.org/static/data/6xhu/6xhu_refined.mtz | 24.70% | Placed the coordinates in a standardized location in the unit cell using ACHESYM. | The coordinates are NOT in a standardized location in the unit cell. | 3C-like proteinase | 46.6 | minimal | Yes | Yes | - | 38.7 | 44.2 | 85.6 | - | ||||||
| 6xip | 1.50 Å | 2020-07-01 | X-ray | - | SARS-CoV-2 | NSP7/NSP8 | Protein-protein complex | 16.07% | 19.91% | 5.80% | - | HKL-3000 | 97.70% | 30.2 | APS (19-ID) | PHENIX | 2020-06-20 | 34197805 | - | - | - | - | N/A | - | - | Non-structural protein 7; Non-structural protein 8 | 64.2 | - | No | - | 82.3 | 38.7 | 82.3 | - | |||||||
| 6z97 | 3.40 Å | 2020-07-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-06-03 | 32585135 | - | - | - | - | N/A | - | - | Spike glycoprotein,Fibritin | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6zco | 1.36 Å | 2020-07-01 | X-ray | - | SARS-CoV-2 | Nucleocapsid | No functional ligands | 14.95% | 19.62% | N/A | - | XDS | 97.80% | 20.2 | SLS (X06DA) | REFMAC | 2020-06-11 | 33039147 | - | - | - | - | N/A | - | - | Nucleoprotein - Dimerization domain | 64.0 | - | No | - | 84.2 | 26.9 | 91.7 | - | |||||||
| 6zdh | 3.70 Å | 2020-07-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-06-14 | 32737466 | - | - | - | - | N/A | - | - | Spike glycoprotein; EY6A heavy chain; EY6A light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6zge | 2.60 Å | 2020-07-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-06-18 | 32647346 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6zgf | 3.10 Å | 2020-07-01 | Cryo-EM | NAG | Bat-CoV-RaTG13 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-06-18 | 32647346 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6zgg | 3.80 Å | 2020-07-01 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2020-06-18 | 32647346 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6zgh | 6.80 Å | 2020-07-01 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2020-06-18 | 32647346 | - | - | - | - | N/A | - | - | Receptor-type tyrosine-protein phosphatase S,Spike glycoprotein,Fibritin | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6zgi | 2.90 Å | 2020-07-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-06-18 | 32647346 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7c2l | 3.10 Å | 2020-07-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-05-08 | 32571838 | - | - | - | - | N/A | - | - | Spike glycoprotein; heavy chain of 4A8; light chain of 4A8 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6xc2 | 3.11 Å | 2020-07-08 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 21.34% | 26.74% | 16.00% | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 88.30% | 7.4 | SSRL (BL12-1) | PHENIX | 2020-06-08 | 32661058 | - | - | - | - | N/A | - | - | Spike protein S1; CC12.1 light chain; CC12.1 heavy chain | 25.9 | - | No | - | 21.3 | 69.4 | 36.6 | - | |||||||
| 6xc3 | 2.70 Å | 2020-07-08 | X-ray | - | SARS-CoV-2 | Spike | Protein-protein complex | 17.73% | 22.53% | 16.70% | - | HKL-2000 | 99.90% | 5.0 | SSRL (BL12-1) | PHENIX | 2020-06-08 | 32661058 | - | - | - | - | N/A | - | - | CR3022 heavy chain; CR3022 light chain; CC12.1 light chain; CC12.1 heavy chain; Spike protein S1 | 63.1 | - | No | - | 59.9 | 84.3 | 56.9 | - | |||||||
| 6xc4 | 2.34 Å | 2020-07-08 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 18.40% | 21.92% | 14.30% | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 98.70% | 10.2 | SSRL (BL12-1) | PHENIX | 2020-06-08 | 32661058 | - | - | - | - | N/A | - | - | Spike protein S1; CC12.3 heavy chain; CC12.3 light chain | 46.1 | - | No | - | 65.8 | 37.7 | 64.1 | - | |||||||
| 6xc7 | 2.88 Å | 2020-07-08 | X-ray | - | SARS-CoV-2 | Spike | Protein-protein complex | 21.89% | 25.91% | 12.90% | - | HKL-2000 | 99.90% | 5.7 | SSRL (BL12-1) | PHENIX | 2020-06-08 | 32661058 | - | - | - | - | N/A | - | - | Spike protein S1; CR3022 heavy chain; CR3022 light chain; CC12.3 heavy chain; CC12.3 light chain | 28.7 | - | No | - | 27.2 | 65.0 | 40.9 | - | |||||||
| 6xhl | 1.47 Å | 2020-07-08 | X-ray | V2M | SARS-CoV | NSP5 (3CLpro) | Functional ligand | 20.35% | 22.29% | 4.70% | N-[(2S)-1-({(2S)-4-hydroxy-3-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | N-[(2S)-1-({(2S)-4-hydroxy-3-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | autoPROC | 74.20% | 12.6 | APS (17-ID) | BUSTER | 2020-06-19 | 33054210 | - | - | - | - | N/A | - | - | 3C-like proteinase | 43.4 | - | No | - | 62.4 | 34.6 | 65.5 | - | ||||||
| 6xhm | 1.41 Å | 2020-07-08 | X-ray | V2M | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.08% | 20.96% | 4.70% | N-[(2S)-1-({(2S)-4-hydroxy-3-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | N-[(2S)-1-({(2S)-4-hydroxy-3-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | autoPROC | 75.00% | 12.3 | APS (17-ID) | BUSTER | 2020-06-19 | 33054210 | - | - | http://covid19.bioreproducibility.org/static/data/6xhm/6xhm_refined.pdb | http://covid19.bioreproducibility.org/static/data/6xhm/6xhm_refined.mtz | 19.20% | Added 2 residues at the C terminus of chain A and 15 waters. Fixed a number of fairly crazy problems with residue occupancies and modified residues A: 41, 106, 110, 125, 139, 222, B: 6, 22, 86, 108, 117, 121, 123, 125. Achesymed and refined with anisotropic B factors. | Missing residues at the C terminus, occupancy problems, coordinates not in standardized location. | 3C-like proteinase | 69.0 | minimal | Yes | Yes | - | 74.4 | 48.4 | 90.7 | - | |||||
| 6xhn | 1.38 Å | 2020-07-08 | X-ray | V3D | SARS-CoV | NSP5 (3CLpro) | Functional ligand | 22.20% | 24.67% | 5.70% | (3S)-3-{[N-(4-methoxy-1H-indole-2-carbonyl)-L-leucyl]amino}-2-oxo-4-[(3S)-2-oxopyrrolidin-3-yl]butyl 2-cyanobenzoate | (3S)-3-{[N-(4-methoxy-1H-indole-2-carbonyl)-L-leucyl]amino}-2-oxo-4-[(3S)-2-oxopyrrolidin-3-yl]butyl 2-cyanobenzoate | autoPROC | 77.90% | 11.6 | APS (17-ID) | BUSTER | 2020-06-19 | 33054210 | - | - | - | - | N/A | - | - | 3C-like proteinase | 24.9 | - | No | - | 38.5 | 15.7 | 71.0 | - | ||||||
| 6xho | 1.45 Å | 2020-07-08 | X-ray | V34 | SARS-CoV | NSP5 (3CLpro) | Functional ligand | 21.09% | 23.87% | 4.70% | ethyl (2E,4S)-4-{[N-(4-methoxy-1H-indole-2-carbonyl)-L-leucyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | ethyl (2E,4S)-4-{[N-(4-methoxy-1H-indole-2-carbonyl)-L-leucyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | autoPROC | 67.00% | 11.5 | APS (17-ID) | BUSTER | 2020-06-19 | 33054210 | - | - | - | - | N/A | - | - | 3C-like proteinase | 30.6 | - | No | - | 46.2 | 25.7 | 65.2 | - | ||||||
| 6xkf | 1.80 Å | 2020-07-08 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 18.98% | 23.90% | 8.90% | - | HKL-3000 | 96.50% | 19.0 | APS (19-ID) | PHENIX | 2020-06-26 | - | - | - | http://covid19.bioreproducibility.org/static/data/6xkf/6xkf_refined.pdb | http://covid19.bioreproducibility.org/static/data/6xkf/6xkf_refined.mtz | 24.50% | Placed the coordinates in a standardized location in the unit cell using ACHESYM. Added TLS. | Coordinates not in standardized location. | 3C-like proteinase | 56.8 | minimal | Yes | Yes | - | - | 45.7 | 76.2 | 66.2 | - | |||||
| 6xkh | 1.28 Å | 2020-07-08 | X-ray | FMT | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 15.63% | 17.49% | 5.90% | FORMIC ACID | FORMIC ACID | HKL-3000 | 98.60% | 40.75 | APS (19-ID) | PHENIX | 2020-06-26 | - | - | - | http://covid19.bioreproducibility.org/static/data/6xkh/6xkh_refined.pdb | http://covid19.bioreproducibility.org/static/data/6xkh/6xkh_refined.mtz | 17.20% | Placed the coordinates in a standardized location in the unit cell using ACHESYM. Deleted EDO401, EDO402. Substituted FMT403 > HOH740, FMT404 > HOH741, FMT405 > HOH742, FMT406 > HOH743, ACT407 > HOH744. Fixed rotamers: His41, Csd145. Added double conformation to Val86, Lys100, Val104, Gln110, Val125, Cys128, Phe134, Arg188, Asn238, Ser267, Glu288. | Coordinates not in standardized location, incorrect rotamers, missing doble conformations. | 3C-like proteinase | 60.3 | moderate | Yes | Yes | - | - | 93.9 | 22.9 | 78.4 | - | ||||
| 6xkm | 2.25 Å | 2020-07-08 | X-ray | SAM | SARS-CoV-2 | NSP10/NSP16 | Functional ligand | 20.65% | 21.26% | N/A | S-ADENOSYLMETHIONINE | S-ADENOSYLMETHIONINE | DIALS | 100.00% | 2.68 | APS (19-ID) | REFMAC | 2020-06-26 | 33972410 | ZN | - | - | - | N/A | - | - | 2'-O-methyltransferase; Non-structural protein 10 | 84.3 | - | No | - | 71.9 | 94.4 | 95.0 | - | ||||||
| 6xmk | 1.70 Å | 2020-07-08 | X-ray | QYS, PG4 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.91% | 21.16% | 7.70% | (1S,2S)-2-[(N-{[(4,4-difluorocyclohexyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-[(N-{[(4,4-difluorocyclohexyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; TETRAETHYLENE GLYCOL | XDS | 98.20% | 8.6 | APS (17-ID) | PHENIX | 2020-06-30 | 32747425 | - | - | http://covid19.bioreproducibility.org/static/data/6xmk/6xmk_refined.pdb | http://covid19.bioreproducibility.org/static/data/6xmk/6xmk_refined.mtz | 21.40% | Achesymed. Added Phe3 to chain A. Added side chains to residues 47, 49, 74 in chain A. Added side chains to residues 47, 49, 154 in chain B. Added 12 waters, deleted water A623 (now part of Tyr154). Put TLS groups as each chain+inhibitor. | Missing sidechains, non-standardized coordinates. | 3C-like proteinase - Full Length | 70.9 | minimal | Yes | Yes | - | 72.7 | 55.4 | 89.6 | - | |||||
| 6zfo | 4.40 Å | 2020-07-08 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-06-17 | 32737466 | - | - | - | - | N/A | - | - | Spike glycoprotein; EY6A heavy chain; EY6A light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6xkl | 3.21 Å | 2020-07-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-06-26 | 32703906 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6xoa | 2.10 Å | 2020-07-15 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 20.55% | 25.09% | 8.80% | - | HKL-3000 | 90.20% | 17.4 | APS (19-ID) | PHENIX | 2020-07-06 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 53.8 | - | No | - | - | 34.5 | 82.4 | 65.4 | - | ||||||
| 6xey | 3.25 Å | 2020-07-22 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-06-14 | 32698192 | - | - | - | - | N/A | - | - | SARS-CoV-2 spike glycoprotein; 2-4 Heavy Chain; 2-4 Light Chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6xqs | 1.90 Å | 2020-07-22 | X-ray | SV6 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.10% | 20.39% | N/A | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide | CrysalisPro | 97.30% | 16.3 | ROTATING ANODE () | PHENIX | 2020-07-10 | 33152262 | - | - | http://covid19.bioreproducibility.org/static/data/6xqs/6xqs_refined.pdb | http://covid19.bioreproducibility.org/static/data/6xqs/6xqs_refined.mtz | 21.10% | Placed the coordinates in a standardized location in the unit cell using ACHESYM. | The coordinates are NOT in a standardized location in the unit cell. | 3C-like proteinase | 60.5 | minimal | Yes | Yes | - | 79.0 | 45.7 | 70.8 | - | |||||
| 6xqt | 2.30 Å | 2020-07-22 | X-ray | NNA | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 22.54% | 27.69% | N/A | (1R,2S,5S)-3-[N-({1-[(tert-butylsulfonyl)methyl]cyclohexyl}carbamoyl)-3-methyl-L-valyl]-N-{(1S)-1-[(1R)-2-(cyclopropyla mino)-1-hydroxy-2-oxoethyl]pentyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-3-[N-({1-[(tert-butylsulfonyl)methyl]cyclohexyl}carbamoyl)-3-methyl-L-valyl]-N-{(1S)-1-[(1R)-2-(cyclopropyla mino)-1-hydroxy-2-oxoethyl]pentyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | CrysalisPro | 96.20% | 5.6 | ROTATING ANODE () | PHENIX | 2020-07-10 | 33152262 | - | - | - | - | N/A | - | - | 3C-like proteinase | 13.8 | - | No | - | 15.7 | 22.0 | 61.8 | - | ||||||
| 6xqu | 2.20 Å | 2020-07-22 | X-ray | U5G | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.60% | 23.38% | N/A | boceprevir (bound form) | boceprevir (bound form) | CrysalisPro | 99.90% | 9.2 | ROTATING ANODE () | PHENIX | 2020-07-10 | 33152262 | - | - | http://covid19.bioreproducibility.org/static/data/6xqu/6xqu_refined.pdb | http://covid19.bioreproducibility.org/static/data/6xqu/6xqu_refined.mtz | 24.76% | Reindexed from I2 to C2 (-h-l,k,h); transformed by ACHESYM. | Non-standard space group, while standard C2 would make it compatible with many other structures of this protein. | 3C-like proteinase | 45.3 | minimal | Yes | Yes | - | 51.3 | 42.8 | 72.0 | - | |||||
| 6xr8 | 2.90 Å | 2020-07-22 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-07-11 | 32694201 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6xra | 3.00 Å | 2020-07-22 | Cryo-EM | NAG, MAN | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; alpha-D-mannopyranose | - | - | () | PHENIX | 2020-07-11 | 32694201 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6xs6 | 3.70 Å | 2020-07-22 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | PHENIX | 2020-07-15 | 32991842 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6zoj | 2.80 Å | 2020-07-22 | Cryo-EM | - | SARS-CoV-2 | NSP1 | No functional ligands | N/A | N/A | N/A | - | - | - | () | PHENIX | 2020-07-07 | 32908316 | MG; ZN | - | - | - | N/A | - | - | 18S ribosomal RNA; 40S ribosomal protein S8; 40S ribosomal protein S9; 40S ribosomal protein S10; 40S ribosomal protein S11; 40S ribosomal protein S12; 40S ribosomal protein S13; 40S ribosomal protein S14; 40S ribosomal protein S15; 40S ribosomal protein S16; 40S ribosomal protein S17; 40S ribosomal protein SA; 40S ribosomal protein S18; 40S ribosomal protein S19; 40S ribosomal protein S20; 40S ribosomal protein S21; 40S ribosomal protein S15a; 40S ribosomal protein S23; 40S ribosomal protein S24; 40S ribosomal protein S25; 40S ribosomal protein S26; 40S ribosomal protein S27; 40S ribosomal protein S3a; 40S ribosomal protein S28; 40S ribosomal protein S29; 40S ribosomal protein S30; Ribosomal protein S27a; Receptor of activated protein C kinase 1; Non-structural protein 1; 60S ribosomal protein L41; 40S ribosomal protein S2; 40S ribosomal protein S3; 40S ribosomal protein S4, X isoform; 40S ribosomal protein S5; 40S ribosomal protein S6; 40S ribosomal protein S7 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6zox | 3.00 Å | 2020-07-22 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-07-08 | 32737467 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6zoy | 3.10 Å | 2020-07-22 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-07-08 | 32737467 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6zoz | 3.50 Å | 2020-07-22 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-07-08 | 32737467 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6zp0 | 3.00 Å | 2020-07-22 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-07-08 | 32737467 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6zp1 | 3.30 Å | 2020-07-22 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-07-08 | 32737467 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6zp2 | 3.10 Å | 2020-07-22 | Cryo-EM | NAG, EIC | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | LINOLEIC ACID | 2-acetamido-2-deoxy-beta-D-glucopyranose; LINOLEIC ACID | - | - | () | PHENIX | 2020-07-08 | 32737467 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 6xez | 3.50 Å | 2020-07-29 | Cryo-EM | 1N7, AF3, ADP | SARS-CoV-2 | NSP7/NSP8/NSP12/NSP13/RNA | Protein-protein complex | N/A | N/A | N/A | CHAPSO; Product RNA; Template RNA; ADENOSINE-5'-DIPHOSPHATE; ALUMINUM FLUORIDE | CHAPSO; ALUMINUM FLUORIDE; ADENOSINE-5'-DIPHOSPHATE | - | - | () | PHENIX | 2020-06-14 | 32783916 | ZN; MG | - | - | - | N/A | - | - | nsp12 - UNP residues 4393-5324; nsp8 - UNP residues 3943-4140; nsp7 - UNP residues 3860-3942; nsp13 - UNP residues 5325-5925; Product RNA; Template RNA | N/A | - | No | - | N/A | N/A | N/A | Product RNA; Template RNA | - | ||||||
| 6xm5 | 3.10 Å | 2020-07-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-06-29 | 33271067 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6xqb | 3.40 Å | 2020-07-29 | Cryo-EM | - | SARS-CoV-2 | NSP7/NSP8/NSP12/RNA | Protein-protein complex | N/A | N/A | N/A | RNA (5'-R(*GP*UP*GP*GP*GP*CP*CP*CP*A)-3') | - | - | - | () | 2020-07-09 | - | ZN; MG | - | - | - | N/A | - | - | RNA-directed RNA polymerase; Non-structural protein 8; Non-structural protein 7; RNA (5'-R(*GP*UP*GP*GP*GP*CP*CP*CP*A)-3') | N/A | - | No | - | - | N/A | N/A | N/A | RNA (5'-R(*GP*UP*GP*GP*GP*CP*CP*CP*A)-3') | - | ||||||
| 6zbp | 1.85 Å | 2020-07-29 | X-ray | NAG | SARS-CoV-2 | Spike | No functional ligands | 18.50% | 21.69% | 8.60% | 2-acetamido-2-deoxy-beta-D-glucopyranose | XSCALE | 99.50% | 18.2 | Diamond (I03) | REFMAC | 2020-06-08 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; H11-H4 | 80.6 | - | No | - | - | 68.2 | 89.2 | 87.6 | - | ||||||
| 6zdg | 4.70 Å | 2020-07-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-06-14 | 32737466 | - | - | - | - | N/A | - | - | Spike glycoprotein; EY6A heavy chain; EY6A light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6zhd | 3.70 Å | 2020-07-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 28.49% | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | REFMAC | 2020-06-22 | - | - | - | - | - | N/A | - | - | Spike glycoprotein,Fibritin; Nanobody H11-H4 | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 6zlw | 2.60 Å | 2020-07-29 | Cryo-EM | - | SARS-CoV-2 | NSP1 | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2020-07-01 | 32680882 | ZN | - | - | - | N/A | - | - | 40S ribosomal protein SA; 40S ribosomal protein S5; 40S ribosomal protein S11; 40S ribosomal protein S10; 40S ribosomal protein S13; 40S ribosomal protein S12; 40S ribosomal protein S14; 40S ribosomal protein S15; 40S ribosomal protein S16; 40S ribosomal protein S17; 40S ribosomal protein S18; 40S ribosomal protein S3a; 40S ribosomal protein S19; 40S ribosomal protein S20; 40S ribosomal protein S15a; 40S ribosomal protein S23; 40S ribosomal protein S24; 40S ribosomal protein S21; 40S ribosomal protein S25; 40S ribosomal protein S27; 40S ribosomal protein S26; 40S ribosomal protein S28; 40S ribosomal protein S2; 40S ribosomal protein S30; 40S ribosomal protein S29; Ubiquitin-40S ribosomal protein S27a; 60S ribosomal protein L41; Receptor of activated protein C kinase 1; 18S ribosomal RNA; Non-structural protein 1; 40S ribosomal protein S4, X isoform; 40S ribosomal protein S3; 40S ribosomal protein S6; 40S ribosomal protein S7; 40S ribosomal protein S8; 40S ribosomal protein S9 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6zm7 | 2.70 Å | 2020-07-29 | Cryo-EM | - | SARS-CoV-2 | NSP1 | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2020-07-01 | 32680882 | ZN; MG | - | - | - | N/A | - | - | 28S ribosomal RNA; 60S ribosomal protein L7a; 60S ribosomal protein L9; 60S ribosomal protein L10-like; 60S ribosomal protein L11; 60S ribosomal protein L13; 60S ribosomal protein L14; 60S ribosomal protein L15; 60S ribosomal protein L13a; 60S ribosomal protein L17; 60S ribosomal protein L18; 5S ribosomal RNA; 60S ribosomal protein L19; 60S ribosomal protein L18a; 60S ribosomal protein L21; 60S ribosomal protein L22; 60S ribosomal protein L23; 60S ribosomal protein L24; 60S ribosomal protein L23a; 60S ribosomal protein L26; 60S ribosomal protein L27; 60S ribosomal protein L27a; 5.8S ribosomal RNA; 60S ribosomal protein L29; 60S ribosomal protein L30; 60S ribosomal protein L31; 60S ribosomal protein L32; 60S ribosomal protein L35a; 60S ribosomal protein L34; 60S ribosomal protein L35; 60S ribosomal protein L36; 60S ribosomal protein L37; 60S ribosomal protein L38; 60S ribosomal protein L8; 60S ribosomal protein L39; Ubiquitin-60S ribosomal protein L40; 60S ribosomal protein L41; 60S ribosomal protein L36a; 60S ribosomal protein L37a; 60S ribosomal protein L28; 60S acidic ribosomal protein P0; 60S ribosomal protein L12; 60S ribosomal protein L10a; 18S ribosomal RNA; 60S ribosomal protein L3; 40S ribosomal protein SA; 40S ribosomal protein S3a; 40S ribosomal protein S3; 40S ribosomal protein S4, X isoform; 40S ribosomal protein S5; 40S ribosomal protein S7; 40S ribosomal protein S8; 40S ribosomal protein S10; 40S ribosomal protein S11; 40S ribosomal protein S15; 60S ribosomal protein L4; 40S ribosomal protein S16; 40S ribosomal protein S17; 40S ribosomal protein S18; 40S ribosomal protein S19; 40S ribosomal protein S20; 40S ribosomal protein S21; 40S ribosomal protein S23; 40S ribosomal protein S26; 40S ribosomal protein S28; 40S ribosomal protein S29; 60S ribosomal protein L5; Receptor of activated protein C kinase 1; 40S ribosomal protein S2; 40S ribosomal protein S6; 40S ribosomal protein S9; 40S ribosomal protein S12; 40S ribosomal protein S13; 40S ribosomal protein S14; 40S ribosomal protein S15a; 40S ribosomal protein S24; 40S ribosomal protein S25; 60S ribosomal protein L6; 40S ribosomal protein S27; 40S ribosomal protein S30; Ubiquitin-40S ribosomal protein S27a; Proliferation-associated protein 2G4; tRNA; Coiled-coil domain-containing protein 124; Non-structural protein 1; 60S ribosomal protein L7 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6zn5 | 3.20 Å | 2020-07-29 | Cryo-EM | - | SARS-CoV-2 | NSP1 | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2020-07-06 | 32680882 | ZN | - | - | - | N/A | - | - | 40S ribosomal protein SA; 40S ribosomal protein S5; 40S ribosomal protein S11; 40S ribosomal protein S10; 40S ribosomal protein S13; 40S ribosomal protein S12; 40S ribosomal protein S14; 40S ribosomal protein S15; 40S ribosomal protein S16; 40S ribosomal protein S17; 40S ribosomal protein S18; 40S ribosomal protein S3a; 40S ribosomal protein S19; 40S ribosomal protein S20; 40S ribosomal protein S15a; 40S ribosomal protein S23; 40S ribosomal protein S24; 40S ribosomal protein S21; 40S ribosomal protein S25; 40S ribosomal protein S27; 40S ribosomal protein S26; 40S ribosomal protein S28; 40S ribosomal protein S2; 40S ribosomal protein S30; 40S ribosomal protein S29; Ribosomal protein S27a; Receptor of activated protein C kinase 1; 18S ribosomal RNA; Non-structural protein 1; Pre-rRNA-processing protein TSR1 homolog; 40S ribosomal protein S4, X isoform; 40S ribosomal protein S3; 40S ribosomal protein S6; 40S ribosomal protein S7; 40S ribosomal protein S8; 40S ribosomal protein S9 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6zok | 2.80 Å | 2020-07-29 | Cryo-EM | - | SARS-CoV-2 | NSP1 | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | PHENIX | 2020-07-07 | 32908316 | ZN; MG | - | - | - | N/A | - | - | 18S ribosomal RNA; 40S ribosomal protein S11; 40S ribosomal protein S13; 40S ribosomal protein S14; 40S ribosomal protein S17; 40S ribosomal protein S21; 40S ribosomal protein S15a; 40S ribosomal protein S23; 40S ribosomal protein S24; 40S ribosomal protein S26; 40S ribosomal protein S27; 40S ribosomal protein SA; 40S ribosomal protein S30; Non-structural protein 1; 60S ribosomal protein L41; 40S ribosomal protein S3a; 40S ribosomal protein S2; 40S ribosomal protein S4, X isoform; 40S ribosomal protein S6; 40S ribosomal protein S7; 40S ribosomal protein S8; 40S ribosomal protein S9 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6zon | 3.00 Å | 2020-07-29 | Cryo-EM | - | SARS-CoV-2 | NSP1 | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2020-07-07 | 32680882 | ZN | - | - | - | N/A | - | - | 40S ribosomal protein SA; 40S ribosomal protein S9; 40S ribosomal protein S11; 40S ribosomal protein S13; 40S ribosomal protein S14; 40S ribosomal protein S21; 40S ribosomal protein S15a; 40S ribosomal protein S23; 40S ribosomal protein S24; 40S ribosomal protein S27; 40S ribosomal protein S30; 40S ribosomal protein S3a; 18S ribosomal RNA; 40S ribosomal protein S17; 40S ribosomal protein S16; 40S ribosomal protein S3; 40S ribosomal protein S5; 40S ribosomal protein S10; 40S ribosomal protein S12; 40S ribosomal protein S15; 40S ribosomal protein S18; 40S ribosomal protein S19; 40S ribosomal protein S2; 40S ribosomal protein S20; 40S ribosomal protein S25; 40S ribosomal protein S28; 40S ribosomal protein S29; Ubiquitin-40S ribosomal protein S27a; Receptor of activated protein C kinase 1; Eukaryotic translation initiation factor 3 subunit I; Eukaryotic translation initiation factor 3 subunit B; Eukaryotic translation initiation factor 3 subunit A,Eukaryotic translation initiation factor 3 subunit A; Eukaryotic translation initiation factor 3 subunit C; 40S ribosomal protein S26; Eukaryotic translation initiation factor 3 subunit E; Eukaryotic translation initiation factor 3 subunit F; Eukaryotic translation initiation factor 3 subunit H; Eukaryotic translation initiation factor 3 subunit K; Eukaryotic translation initiation factor 3 subunit L; Eukaryotic translation initiation factor 3 subunit M; Eukaryotic translation initiation factor 3 subunit D; Unknown factor; Host translation inhibitor Nsp1; 40S ribosomal protein S4, X isoform; 60S ribosomal protein L41; 40S ribosomal protein S6; 40S ribosomal protein S7; 40S ribosomal protein S8 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6zow | 3.00 Å | 2020-07-29 | Cryo-EM | NAG, MAN | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; alpha-D-mannopyranose | - | - | () | 2020-07-07 | 33063791 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6zp4 | 2.90 Å | 2020-07-29 | Cryo-EM | GTP | SARS-CoV-2 | NSP1 | Protein-protein complex | N/A | N/A | N/A | GUANOSINE-5'-TRIPHOSPHATE | GUANOSINE-5'-TRIPHOSPHATE | - | - | () | 2020-07-08 | 32680882 | MG; ZN | - | - | - | N/A | - | - | 40S ribosomal protein SA; 40S ribosomal protein S9; 40S ribosomal protein S11; 40S ribosomal protein S13; 40S ribosomal protein S21; 40S ribosomal protein S15a; 40S ribosomal protein S24; 40S ribosomal protein S27; 40S ribosomal protein S30; 18S ribosomal RNA; 40S ribosomal protein S17; 40S ribosomal protein S3a; 40S ribosomal protein S3; 40S ribosomal protein S5; 40S ribosomal protein S10; 40S ribosomal protein S12; 40S ribosomal protein S15; 40S ribosomal protein S16; 40S ribosomal protein S18; 40S ribosomal protein S19; 40S ribosomal protein S20; 40S ribosomal protein S25; 40S ribosomal protein S2; 40S ribosomal protein S28; 40S ribosomal protein S29; Ubiquitin-40S ribosomal protein S27a; Receptor of activated protein C kinase 1; 40S ribosomal protein S14; 40S ribosomal protein S23; Eukaryotic translation initiation factor 1A, X-chromosomal; Eukaryotic translation initiation factor 3 subunit I; Eukaryotic translation initiation factor 3 subunit B; Eukaryotic translation initiation factor 3 subunit A; 40S ribosomal protein S26; Eukaryotic translation initiation factor 3 subunit C; Eukaryotic translation initiation factor 3 subunit E; Eukaryotic translation initiation factor 3 subunit F; Eukaryotic translation initiation factor 3 subunit H; Eukaryotic translation initiation factor 3 subunit K; Eukaryotic translation initiation factor 3 subunit L; Eukaryotic translation initiation factor 3 subunit M; Eukaryotic translation initiation factor 3 subunit D; Unknown factor; tRNA; 40S ribosomal protein S4, X isoform; Eukaryotic translation initiation factor 2 subunit 2; Eukaryotic translation initiation factor 2 subunit 1; Eukaryotic translation initiation factor 2 subunit 3; Eukaryotic translation initiation factor 1; Non-structural protein 1; 60S ribosomal protein L41; 40S ribosomal protein S6; 40S ribosomal protein S7; 40S ribosomal protein S8 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6zp5 | 3.10 Å | 2020-07-29 | Cryo-EM | NAG, MAN | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; alpha-D-mannopyranose | - | - | () | 2020-07-08 | 33063791 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6zp7 | 3.30 Å | 2020-07-29 | Cryo-EM | NAG, MAN | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; alpha-D-mannopyranose | - | - | () | 2020-07-08 | 33063791 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6zsl | 1.94 Å | 2020-07-29 | X-ray | PO4 | SARS-CoV-2 | NSP13 | No functional ligands | 20.94% | 25.31% | 5.30% | PHOSPHATE ION | PHOSPHATE ION | XDS | 97.30% | 11.1 | Diamond (I04-1) | PHENIX | 2020-07-15 | 34381037 | ZN | - | - | - | N/A | - | - | SARS-CoV-2 helicase NSP13 | 20.2 | - | No | - | 32.4 | 26.0 | 56.5 | - | ||||||
| 7c7p | 1.74 Å | 2020-07-29 | X-ray | SV6, FK3 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.42% | 21.59% | 9.80% | (3~{S},3~{a}~{S},6~{a}~{R})-~{N}-[(2~{R},3~{S})-1-(cyclopropylamino)-2-oxidanyl-1-oxidanylidene-hexan-3-yl]-2-methanoyl-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide; (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide; (3~{S},3~{a}~{S},6~{a}~{R})-~{N}-[(2~{R},3~{S})-1-(cyclopropylamino)-2-oxidanyl-1-oxidanylidene-hexan-3-yl]-2-methanoyl-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide | XDS | 99.90% | 22.8 | SSRF (BL19U1) | REFMAC | 2020-05-26 | 33602867 | - | - | http://covid19.bioreproducibility.org/static/data/7c7p/7c7p_hkl_refine_23_tls.pdb | http://covid19.bioreproducibility.org/static/data/7c7p/7c7p_hkl_refine_23_coot.mtz | 19.30% | Standardized with Achesym server; added TLS groups; added missing waters; chaned coformation of the ligand in chain B and changed its occupancy to 0.6; changed rotamers for A5 A47 A48 A50 B58 B60 B67 A67 B76 A82 A86 A/B87 B82 A90 A100 B110 A/B154 A155 B188 A216 A222 B262 B269 B268 B298 A196 B100 | Not in the standard position in the unit cell, flipped sidechains, incorrect rotamers, missing waters | main protease | 36.3 | moderate | Yes | Yes | - | 69.1 | 32.6 | 47.1 | http://covid19.bioreproducibility.org/static/data/7c7p/7c7p_hkl_refine_23_7C7P_crystal1_report.pdf | |||||
| 7jfq | 1.55 Å | 2020-07-29 | X-ray | FMT | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 16.91% | 19.81% | 9.90% | FORMIC ACID | FORMIC ACID | HKL-3000 | 99.20% | 21.8 | APS (19-ID) | PHENIX | 2020-07-17 | - | - | - | http://covid19.bioreproducibility.org/static/data/7jfq/7jfq_refined.pdb | http://covid19.bioreproducibility.org/static/data/7jfq/7jfq_refined.mtz | 19.70% | Placed the coordinates in a standardized location in the unit cell using ACHESYM. Added TLS. | Coordinates not in standardized location. | 3C-like proteinase | 61.5 | minimal | Yes | Yes | - | - | 82.9 | 39.9 | 74.7 | - | ||||
| 6wc1 | 2.40 Å | 2020-08-05 | X-ray | - | SARS-CoV-2 | NSP9 | No functional ligands | 23.20% | 27.80% | N/A | - | MOSFLM | 94.80% | 23.4 | ROTATING ANODE () | PHENIX | 2020-03-28 | - | - | - | http://covid19.bioreproducibility.org/static/data/6wc1/6wc1_rerefined.pdb | http://covid19.bioreproducibility.org/static/data/6wc1/6wc1_rerefined.mtz | 26.74% | Standardized with Achesym server; added TLS groups; added missing waters; THR34-ARG39 remodelling; flipped ASN98 A sidechain; changed rotamers for VAL110, THR24 A, LYS92 B, LYS52 A, ARG74 A, THR79 B, THR64 A, ILE92 A, LEU106 A, LEU112 A, ASP60 A and B THR34 A. There seems to be something bigger next to ARG 39 A, but the density is too small to fir CIT, modeled 3 waters. | Not in the standard position in the unit cell, flipped sidechains, incorrect rotamers, missing waters | SARS-coV-2 Non-structural protein 9 | 8.4 | moderate | Yes | Yes | - | - | 15.0 | 23.8 | 44.3 | - | |||||
| 6wco | 1.48 Å | 2020-08-05 | X-ray | MES, X47 | SARS CoV Urbani | NSP5 (3CLpro) | Functional ligand | 16.89% | 19.44% | N/A | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID; N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclopentylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID; N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclopentylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide | HKL-2000 | 99.60% | 39.6 | APS (21-ID-G) | PHENIX | 2020-03-30 | - | - | - | - | - | N/A | - | - | Main protease | 54.2 | - | No | - | - | 85.3 | 45.7 | 52.2 | - | |||||
| 6zwv | 3.50 Å | 2020-08-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-07-28 | 32805734 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7jir | 2.09 Å | 2020-08-05 | X-ray | TTT, MES | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 18.58% | 19.98% | 14.50% | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | HKL-3000 | 99.80% | 7.4 | APS (19-ID) | REFMAC | 2020-07-23 | 33531496 | ZN | - | - | - | N/A | - | - | Papain-like protease | 57.3 | - | No | 81.9 | 23.6 | 83.6 | - | |||||||
| 7jit | 1.95 Å | 2020-08-05 | X-ray | Y95, MES | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 17.49% | 19.02% | 9.70% | 5-[(carbamoylcarbamoyl)amino]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | 5-[(carbamoylcarbamoyl)amino]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | HKL-3000 | 99.50% | 5.9 | APS (19-ID) | REFMAC | 2020-07-23 | 33531496 | ZN | - | - | - | N/A | - | - | Papain-like protease | 65.9 | - | No | 87.6 | 30.4 | 88.8 | - | |||||||
| 7jiv | 2.05 Å | 2020-08-05 | X-ray | VBY, MES | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 18.78% | 20.09% | 10.10% | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID; 5-(acryloylamino)-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide | 5-(acryloylamino)-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | HKL-3000 | 99.90% | 6.0 | APS (19-ID) | REFMAC | 2020-07-23 | 33531496 | ZN | - | - | - | N/A | - | - | Papain-like protease | 38.9 | - | No | 81.2 | 12.8 | 59.9 | - | |||||||
| 7jiw | 2.30 Å | 2020-08-05 | X-ray | VBY, MES | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 21.21% | 23.95% | 9.50% | 5-(acryloylamino)-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | 5-(acryloylamino)-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | HKL-3000 | 99.90% | 6.3 | APS (19-ID) | REFMAC | 2020-07-23 | 33531496 | ZN | - | - | - | N/A | - | - | Papain-Like Protease | 37.8 | - | No | 45.4 | 25.2 | 81.1 | - | |||||||
| 6vgy | 2.05 Å | 2020-08-12 | X-ray | QZJ | MERS-CoV | NSP5 (3CLpro) | Functional ligand | 19.18% | 23.89% | 8.20% | N~2~-{[(trans-4-ethylcyclohexyl)oxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | N~2~-{[(trans-4-ethylcyclohexyl)oxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | XDS | 98.90% | 12.2 | APS (17-ID) | PHENIX | 2020-01-09 | 32747425 | - | - | - | - | N/A | - | - | Orf1a protein | 61.4 | - | No | - | 46.1 | 78.2 | 72.9 | - | ||||||
| 6vgz | 2.25 Å | 2020-08-12 | X-ray | QZG | MERS-CoV | NSP5 (3CLpro) | Functional ligand | 18.65% | 24.79% | 12.70% | N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[trans-4-(propan-2-yl)cyclohexyl]oxy}carbonyl)-L-leucinamide | N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[trans-4-(propan-2-yl)cyclohexyl]oxy}carbonyl)-L-leucinamide | XDS | 99.90% | 9.8 | APS (17-ID) | PHENIX | 2020-01-09 | 32747425 | - | - | - | - | N/A | - | - | Orf1a protein | 57.0 | - | No | - | 37.4 | 75.5 | 75.6 | - | ||||||
| 6vh0 | 1.95 Å | 2020-08-12 | X-ray | QZD | MERS-CoV | NSP5 (3CLpro) | Functional ligand | 19.51% | 24.04% | 9.80% | N~2~-{[(5-ethyl-1,3-dioxan-5-yl)oxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | N~2~-{[(5-ethyl-1,3-dioxan-5-yl)oxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | XDS | 100.00% | 10.3 | APS (17-ID) | PHENIX | 2020-01-09 | 32747425 | - | - | - | - | N/A | - | - | Orf1a protein | 61.0 | - | No | - | 44.5 | 68.1 | 83.8 | - | ||||||
| 6vh1 | 2.30 Å | 2020-08-12 | X-ray | QZ7 | MERS-CoV | NSP5 (3CLpro) | Functional ligand | 19.17% | 25.34% | 6.20% | N~2~-{[(4,4-difluorocyclohexyl)oxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | N~2~-{[(4,4-difluorocyclohexyl)oxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | XDS | 100.00% | 17.8 | APS (17-ID) | PHENIX | 2020-01-09 | 32747425 | - | - | - | - | N/A | - | - | Orf1a protein | - | - | No | - | 32.2 | - | - | - | ||||||
| 6vh2 | 2.26 Å | 2020-08-12 | X-ray | QZ4 | MERS-CoV | NSP5 (3CLpro) | Functional ligand | 18.35% | 24.81% | 12.80% | 4,4-difluorocyclohexyl [(2S)-3-cyclohexyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamate | 4,4-difluorocyclohexyl [(2S)-3-cyclohexyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamate | XDS | 99.60% | 10.3 | APS (17-ID) | PHENIX | 2020-01-09 | 32747425 | - | - | - | - | N/A | - | - | Orf1a protein | 58.4 | - | No | - | 37.0 | 70.8 | 83.6 | - | ||||||
| 6vh3 | 2.20 Å | 2020-08-12 | X-ray | QYS | MERS-CoV | NSP5 (3CLpro) | Functional ligand | 18.30% | 24.53% | 14.10% | (1S,2S)-2-[(N-{[(4,4-difluorocyclohexyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-[(N-{[(4,4-difluorocyclohexyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | XDS | 100.00% | 9.3 | APS (17-ID) | PHENIX | 2020-01-09 | 32747425 | - | - | - | - | N/A | - | - | Orf1a protein | 52.5 | - | No | - | 39.8 | 75.9 | 64.2 | - | ||||||
| 6w2a | 1.65 Å | 2020-08-12 | X-ray | VDJ, QYS | SARS-CoV | NSP5 (3CLpro) | Functional ligand | 17.20% | 20.30% | 6.90% | [4,4-bis(fluoranyl)cyclohexyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | [4,4-bis(fluoranyl)cyclohexyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate; (1S,2S)-2-[(N-{[(4,4-difluorocyclohexyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | XDS | 99.90% | 12.6 | APS (17-ID) | PHENIX | 2020-03-05 | 32747425 | - | - | - | - | N/A | - | - | Replicase polyprotein 1a | 80.3 | - | No | - | 79.6 | 83.5 | 80.7 | - | ||||||
| 6xlu | 2.40 Å | 2020-08-12 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-06-29 | 33271067 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6xm0 | 2.70 Å | 2020-08-12 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-06-29 | 33271067 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6xm3 | 2.90 Å | 2020-08-12 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-06-29 | 33271067 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 6xm4 | 2.90 Å | 2020-08-12 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-06-29 | 33271067 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6zme | 3.00 Å | 2020-08-12 | Cryo-EM | SF4, ADP | SARS-CoV-2 | NSP1 | No functional ligands | N/A | N/A | N/A | IRON/SULFUR CLUSTER; ADENOSINE-5'-DIPHOSPHATE | IRON/SULFUR CLUSTER; ADENOSINE-5'-DIPHOSPHATE | - | - | () | 2020-07-02 | 32680882 | ZN; MG | - | - | - | N/A | - | - | 28S ribosomal RNA; 60S ribosomal protein L7a; 60S ribosomal protein L9; 60S ribosomal protein L10-like; 60S ribosomal protein L11; 60S ribosomal protein L13; 60S ribosomal protein L14; 60S ribosomal protein L15; 60S ribosomal protein L13a; 60S ribosomal protein L17; 60S ribosomal protein L18; 5S ribosomal RNA; 60S ribosomal protein L19; 60S ribosomal protein L18a; 60S ribosomal protein L21; 60S ribosomal protein L22; 60S ribosomal protein L23; 60S ribosomal protein L24; 60S ribosomal protein L23a; 60S ribosomal protein L26; 60S ribosomal protein L27; 60S ribosomal protein L27a; 5.8S ribosomal RNA; 60S ribosomal protein L29; 60S ribosomal protein L30; 60S ribosomal protein L31; 60S ribosomal protein L32; 60S ribosomal protein L35a; 60S ribosomal protein L34; 60S ribosomal protein L35; 60S ribosomal protein L36; 60S ribosomal protein L37; 60S ribosomal protein L38; 60S ribosomal protein L8; 60S ribosomal protein L39; Ubiquitin-60S ribosomal protein L40; 60S ribosomal protein L41; 60S ribosomal protein L36a; 60S ribosomal protein L37a; 60S ribosomal protein L28; 60S acidic ribosomal protein P0; 60S ribosomal protein L12; 60S ribosomal protein L10a; 18S ribosomal RNA; 60S ribosomal protein L3; 40S ribosomal protein SA; 40S ribosomal protein S3a; 40S ribosomal protein S3; 40S ribosomal protein S4, X isoform; 40S ribosomal protein S5; 40S ribosomal protein S7; 40S ribosomal protein S8; 40S ribosomal protein S10; 40S ribosomal protein S11; 40S ribosomal protein S15; 60S ribosomal protein L4; 40S ribosomal protein S16; 40S ribosomal protein S17; 40S ribosomal protein S18; 40S ribosomal protein S19; 40S ribosomal protein S20; 40S ribosomal protein S21; 40S ribosomal protein S23; 40S ribosomal protein S26; 40S ribosomal protein S28; 40S ribosomal protein S29; 60S ribosomal protein L5; Receptor of activated protein C kinase 1; 40S ribosomal protein S2; 40S ribosomal protein S6; 40S ribosomal protein S9; 40S ribosomal protein S12; 40S ribosomal protein S13; 40S ribosomal protein S14; 40S ribosomal protein S15a; 40S ribosomal protein S24; 40S ribosomal protein S25; 60S ribosomal protein L6; 40S ribosomal protein S27; 40S ribosomal protein S30; Ubiquitin-40S ribosomal protein S27a; Proliferation-associated protein 2G4; tRNA; Coiled-coil domain-containing protein 124; Non-structural protein 1; ATP-binding cassette sub-family E member 1; Eukaryotic peptide chain release factor subunit 1; 60S ribosomal protein L7 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6zrt | 2.10 Å | 2020-08-12 | X-ray | SV6 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.88% | 23.66% | 5.00% | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide | XDS | 99.30% | 22.1 | PETRA III, DESY (P11) | REFMAC | 2020-07-14 | 34041486 | - | - | http://covid19.bioreproducibility.org/static/data/6zrt/6zrt_refmac.pdb | http://covid19.bioreproducibility.org/static/data/6zrt/6zrt_refmac.mtz | 23.60% | Standardized with Achesym server and added TLS groups | Well refined structure with a clearly defined ligand electon density | Main Protease | 57.1 | minimal | Yes | Yes | - | 48.3 | 64.3 | 76.1 | - | |||||
| 6zru | 2.10 Å | 2020-08-12 | X-ray | U5G | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.76% | 21.54% | 4.00% | boceprevir (bound form) | boceprevir (bound form) | XDS | 100.00% | 23.9 | PETRA III, DESY (P11) | REFMAC | 2020-07-14 | 34041486 | - | - | http://covid19.bioreproducibility.org/static/data/6zru/6zru_refined.pdb | http://covid19.bioreproducibility.org/static/data/6zru/6zru_refined.mtz | 23.67% | Standardized with Achesym server, changed space group to C2. | Well refined structure with a clearly defined ligand electon density | Main Protease | 63.5 | minimal | Yes | Yes | - | 69.4 | 59.9 | 72.4 | - | |||||
| 7cah | 3.90 Å | 2020-08-12 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | PHENIX | 2020-06-08 | 32703908 | - | - | - | - | N/A | - | - | Light chain of H014 Fab; Heavy chain of H014 Fab; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7jn2 | 1.93 Å | 2020-08-12 | X-ray | Y41, UNX | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 18.58% | 20.89% | 11.50% | 3-amino-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide; UNKNOWN ATOM OR ION | 3-amino-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide; UNKNOWN ATOM OR ION | HKL-3000 | 99.80% | 7.1 | APS (19-ID) | REFMAC | 2020-08-03 | - | ZN | - | http://covid19.bioreproducibility.org/static/data/7jn2/7jn2_rerefinement.pdb | http://covid19.bioreproducibility.org/static/data/7jn2/7jn2_rerefinement.mtz | 19.77% | Standardized with Achesym server; added TLS groups; added many missing water molecules; added VAL2, ARG3; changed rotamer for HIS17; changed UNK to waters next to HIS255; added alternative conformation for HIS272; added partially occupied MES next to TRP106, split TRP106 to alternative conformation. | Not in the standard position in the unit cell, incorrect rotamers, missing waters, missing residues | Papain-Like Protease | 46.6 | moderate | Yes | Yes | - | 75.1 | 15.2 | 78.2 | - | |||||
| 6x79 | 2.90 Å | 2020-08-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-05-29 | 32753755 | - | - | - | - | N/A | - | - | SARS-CoV-2 spike glycoprotein - ectodomain(UNP residues 14-1211) | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6xr3 | 1.45 Å | 2020-08-19 | X-ray | V7G | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 14.34% | 18.74% | 6.28% | N-[(2S)-1-({(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | N-[(2S)-1-({(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | HKL-2000 | 94.00% | 19.17 | APS (21-ID-F) | PHENIX | 2020-07-10 | - | - | - | http://covid19.bioreproducibility.org/static/data/6xr3/6xr3_refined.pdb | http://covid19.bioreproducibility.org/static/data/6xr3/6xr3_refined.mtz | 18.40% | Achesymed. Deleted residues 46-48 (no density). Adjusted residues: 86, 100, 117, 128, 162, 165, 217, 263, 279, 288, 298. Used anisotropic refinement. | Coordinates not in standardized location. Few residues without density. | 3C-like proteinase | 57.3 | minimal | Yes | Yes | - | - | 89.1 | 21.9 | 78.0 | - | ||||
| 6xrz | 6.90 Å | 2020-08-19 | Cryo-EM | - | SARS-CoV-2 | RNA/RNA | No functional ligands | N/A | N/A | N/A | Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | - | - | - | () | 2020-07-14 | 32743589 | - | - | - | - | N/A | - | - | Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome - GB residues 13434-13521 | N/A | - | No | - | N/A | N/A | N/A | Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | - | |||||||
| 6zmi | 2.60 Å | 2020-08-19 | Cryo-EM | - | SARS-CoV-2 | NSP1 | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | 2020-07-02 | 32680882 | ZN; MG | - | - | - | N/A | - | - | 28S ribosomal RNA; 60S ribosomal protein L7a; 60S ribosomal protein L9; 60S ribosomal protein L10-like; 60S ribosomal protein L11; 60S ribosomal protein L13; 60S ribosomal protein L14; 60S ribosomal protein L15; 60S ribosomal protein L13a; 60S ribosomal protein L17; 60S ribosomal protein L18; 5S ribosomal RNA; 60S ribosomal protein L19; 60S ribosomal protein L18a; 60S ribosomal protein L21; 60S ribosomal protein L22; 60S ribosomal protein L23; 60S ribosomal protein L24; 60S ribosomal protein L23a; 60S ribosomal protein L26; 60S ribosomal protein L27; 60S ribosomal protein L27a; 5.8S ribosomal RNA; 60S ribosomal protein L29; 60S ribosomal protein L30; 60S ribosomal protein L31; 60S ribosomal protein L32; 60S ribosomal protein L35a; 60S ribosomal protein L34; 60S ribosomal protein L35; 60S ribosomal protein L36; 60S ribosomal protein L37; 60S ribosomal protein L38; 60S ribosomal protein L8; 60S ribosomal protein L39; Ubiquitin-60S ribosomal protein L40; 60S ribosomal protein L41; 60S ribosomal protein L36a; 60S ribosomal protein L37a; 60S ribosomal protein L28; 60S acidic ribosomal protein P0; 60S ribosomal protein L12; 60S ribosomal protein L10a; 18S ribosomal RNA; 60S ribosomal protein L3; 40S ribosomal protein SA; 40S ribosomal protein S3a; 40S ribosomal protein S3; 40S ribosomal protein S4, X isoform; 40S ribosomal protein S5; 40S ribosomal protein S7; 40S ribosomal protein S8; 40S ribosomal protein S10; 40S ribosomal protein S11; 40S ribosomal protein S15; 60S ribosomal protein L4; 40S ribosomal protein S16; 40S ribosomal protein S17; 40S ribosomal protein S18; 40S ribosomal protein S19; 40S ribosomal protein S20; 40S ribosomal protein S21; 40S ribosomal protein S23; 40S ribosomal protein S26; 40S ribosomal protein S28; 40S ribosomal protein S29; 60S ribosomal protein L5; Receptor of activated protein C kinase 1; 40S ribosomal protein S2; 40S ribosomal protein S6; 40S ribosomal protein S9; 40S ribosomal protein S12; 40S ribosomal protein S13; 40S ribosomal protein S14; 40S ribosomal protein S15a; 40S ribosomal protein S24; 40S ribosomal protein S25; 60S ribosomal protein L6; 40S ribosomal protein S27; 40S ribosomal protein S30; Ubiquitin-40S ribosomal protein S27a; Proliferation-associated protein 2G4; tRNA; Cell growth-regulating nucleolar protein; Non-structural protein 1; 60S ribosomal protein L7 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6zmo | 3.10 Å | 2020-08-19 | Cryo-EM | - | SARS-CoV-2 | NSP1 | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | 2020-07-03 | 32680882 | MG; ZN | - | - | - | N/A | - | - | 28S ribosomal RNA; 60S ribosomal protein L7a; 60S ribosomal protein L9; 60S ribosomal protein L10-like; 60S ribosomal protein L11; 60S ribosomal protein L13; 60S ribosomal protein L14; 60S ribosomal protein L15; 60S ribosomal protein L13a; 60S ribosomal protein L17; 60S ribosomal protein L18; 5S ribosomal RNA; 60S ribosomal protein L19; 60S ribosomal protein L18a; 60S ribosomal protein L21; 60S ribosomal protein L22; 60S ribosomal protein L23; 60S ribosomal protein L24; 60S ribosomal protein L23a; 60S ribosomal protein L26; 60S ribosomal protein L27; 60S ribosomal protein L27a; 5.8S ribosomal RNA; 60S ribosomal protein L29; 60S ribosomal protein L30; 60S ribosomal protein L31; 60S ribosomal protein L32; 60S ribosomal protein L35a; 60S ribosomal protein L34; 60S ribosomal protein L35; 60S ribosomal protein L36; 60S ribosomal protein L37; 60S ribosomal protein L38; 60S ribosomal protein L8; 60S ribosomal protein L39; Ubiquitin-60S ribosomal protein L40; 60S ribosomal protein L41; 60S ribosomal protein L36a; 60S ribosomal protein L37a; 60S ribosomal protein L28; 60S acidic ribosomal protein P0; 60S ribosomal protein L12; 60S ribosomal protein L10a; 18S ribosomal RNA; 60S ribosomal protein L3; 40S ribosomal protein SA; 40S ribosomal protein S3a; 40S ribosomal protein S3; 40S ribosomal protein S4, X isoform; 40S ribosomal protein S5; 40S ribosomal protein S7; 40S ribosomal protein S8; 40S ribosomal protein S10; 40S ribosomal protein S11; 40S ribosomal protein S15; 60S ribosomal protein L4; 40S ribosomal protein S16; 40S ribosomal protein S17; 40S ribosomal protein S18; 40S ribosomal protein S19; 40S ribosomal protein S20; 40S ribosomal protein S21; 40S ribosomal protein S23; 40S ribosomal protein S26; 40S ribosomal protein S28; 40S ribosomal protein S29; 60S ribosomal protein L5; Receptor of activated protein C kinase 1; 40S ribosomal protein S2; 40S ribosomal protein S6; 40S ribosomal protein S9; 40S ribosomal protein S12; 40S ribosomal protein S13; 40S ribosomal protein S14; 40S ribosomal protein S15a; 40S ribosomal protein S24; 40S ribosomal protein S25; 60S ribosomal protein L6; 40S ribosomal protein S27; 40S ribosomal protein S30; Ubiquitin-40S ribosomal protein S27a; Proliferation-associated protein 2G4; tRNA; tRNA; Elongation factor 1-alpha 1; Cell growth-regulating nucleolar protein; Non-structural protein 1; 60S ribosomal protein L7 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6zmt | 3.00 Å | 2020-08-19 | Cryo-EM | - | SARS-CoV-2 | NSP1 | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | 2020-07-03 | 32680882 | ZN | - | - | - | N/A | - | - | 40S ribosomal protein SA; 40S ribosomal protein S5; 40S ribosomal protein S11; 40S ribosomal protein S10; 40S ribosomal protein S13; 40S ribosomal protein S12; 40S ribosomal protein S14; 40S ribosomal protein S15; 40S ribosomal protein S16; 40S ribosomal protein S17; 40S ribosomal protein S18; 40S ribosomal protein S3a; 40S ribosomal protein S19; 40S ribosomal protein S20; 40S ribosomal protein S15a; 40S ribosomal protein S23; 40S ribosomal protein S24; 40S ribosomal protein S21; 40S ribosomal protein S25; 40S ribosomal protein S27; 40S ribosomal protein S28; 40S ribosomal protein S30; 40S ribosomal protein S2; 40S ribosomal protein S29; Ubiquitin-40S ribosomal protein S27a; Receptor of activated protein C kinase 1; 18S ribosomal RNA; Non-structural protein 1; Pre-rRNA-processing protein TSR1 homolog; 40S ribosomal protein S4, X isoform; 40S ribosomal protein S3; 40S ribosomal protein S6; 40S ribosomal protein S7; 40S ribosomal protein S8; 40S ribosomal protein S9 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7com | 2.25 Å | 2020-08-19 | X-ray | U5G | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.78% | 24.63% | 17.70% | boceprevir (bound form) | boceprevir (bound form) | XDS | 99.90% | 11.7 | SSRF (BL19U1) | BUSTER | 2020-08-04 | 33602867 | - | - | http://covid19.bioreproducibility.org/static/data/7com/7com_rerefined.pdb | http://covid19.bioreproducibility.org/static/data/7com/7com_rerefined.mtz | 24.91% | Achesym, added 3 missing residues in chain B (THR 45, LEU 50, ASN 51 B), flipped some sidechains, modeled disorder, refined U5G in chain A. | Missing reidues and flipped sidechain. | main protease | 40.4 | moderate | Yes | Yes | - | 38.8 | 49.8 | 68.2 | - | |||||
| 6m39 | 3.55 Å | 2020-08-26 | Cryo-EM | NAG | SADS-CoV | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-03-03 | 32817223 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 6w79 | 1.46 Å | 2020-08-26 | X-ray | X77, MES | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 15.64% | 17.73% | N/A | N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | HKL-2000 | 99.70% | 41.3 | APS (21-ID-G) | PHENIX | 2020-03-18 | - | - | - | http://covid19.bioreproducibility.org/static/data/6w79/6w79_refined.pdb | http://covid19.bioreproducibility.org/static/data/6w79/6w79_refined.mtz | 15.40% | Achesymed. Minor coordinate adjustments. Used anisotropic refinement. | The coordinates are NOT in a standardized location in the unit cell. | Main protease | 79.2 | minimal | Yes | Yes | - | - | 93.1 | 60.2 | 86.8 | - | ||||
| 7cn9 | 4.70 Å | 2020-08-26 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-07-30 | 32755598 | - | - | - | - | N/A | - | - | Spike protein S1, Spike protein S2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7jhe | 2.25 Å | 2020-08-26 | X-ray | SAH, V9G, MGP | SARS-CoV-2 | NSP10/NSP16 | Functional ligand | 22.31% | 24.85% | N/A | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE; 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE; S-ADENOSYL-L-HOMOCYSTEINE | S-ADENOSYL-L-HOMOCYSTEINE; 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE; 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE | DIALS | 100.00% | 2.45 | APS (19-ID) | REFMAC | 2020-07-20 | 33972410 | ZN | - | - | - | N/A | - | - | 2'-O-methyltransferase; Non-structural protein 10 | 68.4 | - | No | - | 36.7 | 87.7 | 87.7 | - | ||||||
| 7jib | 2.65 Å | 2020-08-26 | X-ray | SAM, SAH, GTA, V9G, MGP | SARS-CoV-2 | NSP10/NSP16 | Functional ligand | 15.49% | 18.09% | 14.80% | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE; S-ADENOSYLMETHIONINE; 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE; 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE; S-ADENOSYL-L-HOMOCYSTEINE | S-ADENOSYLMETHIONINE; S-ADENOSYL-L-HOMOCYSTEINE; P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE; 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE; 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE | HKL-3000 | 99.60% | 15.0 | APS (19-ID) | REFMAC | 2020-07-23 | 33972410 | ZN | - | - | - | N/A | - | - | 2'-O-methyltransferase; Non-structural protein 10 | 69.4 | - | No | - | 91.8 | 25.0 | 97.4 | - | ||||||
| 7jji | 3.60 Å | 2020-08-26 | Cryo-EM | NAG, VCG, EIC | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | LINOLEIC ACID; 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-hydroxyethyl 2-deoxy-3,5-bis-O-(2-hydroxyethyl)-6-O-(2-{[(9E)-octadec-9-enoyl]oxy}ethyl)-alpha-L-xylo-hexofuranoside | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-hydroxyethyl 2-deoxy-3,5-bis-O-(2-hydroxyethyl)-6-O-(2-{[(9E)-octadec-9-enoyl]oxy}ethyl)-alpha-L-xylo-hexofuranoside; LINOLEIC ACID | - | - | () | 2020-07-26 | 33082295 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7jjj | 4.50 Å | 2020-08-26 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-07-26 | 33082295 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7jlt | 2.70 Å | 2020-08-26 | X-ray | - | SARS-CoV-2 | NSP7/NSP8 | No functional ligands | 23.65% | 28.30% | 12.90% | - | HKL-3000 | 99.30% | 4.8 | ALS (5.0.1) | PHENIX | 2020-07-30 | 33999154 | - | - | - | - | N/A | - | - | Non-structural protein 7; Non-structural protein 8 | 29.0 | - | No | - | 12.7 | 90.8 | 30.3 | - | |||||||
| 7jmo | 2.36 Å | 2020-08-26 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 19.51% | 23.81% | 16.30% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 97.20% | 5.7 | SSRL (BL12-1) | PHENIX | 2020-08-02 | 33027617 | - | - | - | - | N/A | - | - | Spike protein S1; COVA2-04 heavy chain; COVA2-04 light chain | 37.3 | - | No | - | 46.7 | 35.8 | 68.1 | - | ||||||
| 7jmp | 1.71 Å | 2020-08-26 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 17.68% | 20.91% | 5.70% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 97.20% | 11.6 | SSRL (BL12-1) | PHENIX | 2020-08-02 | 33027617 | - | - | - | - | N/A | - | - | Spike protein S1; COVA2-39 heavy chain; COVA2-39 light chain | 52.6 | - | No | - | 74.7 | 28.6 | 76.7 | - | ||||||
| 7jpe | 2.18 Å | 2020-08-26 | X-ray | GTA, SAM, 8NK | SARS-CoV-2 | NSP10/NSP16 | Functional ligand | 21.70% | 23.70% | N/A | S-ADENOSYLMETHIONINE; 7-methylguanosine 5'-diphosphate; P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE; S-ADENOSYLMETHIONINE; 7-methylguanosine 5'-diphosphate | DIALS | 100.00% | 2.55 | APS (19-ID) | REFMAC | 2020-08-07 | 33972410 | ZN | - | - | - | N/A | - | - | 2'-O-methyltransferase; Non-structural protein 10 | 75.9 | - | No | - | 47.7 | 87.7 | 94.8 | - | ||||||
| 7jr3 | 1.55 Å | 2020-08-26 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 15.91% | 18.32% | 3.80% | - | XDS | 96.00% | 15.53 | LNLS SIRUS (MANACA) | PHENIX | 2020-08-11 | - | - | - | http://covid19.bioreproducibility.org/static/data/7jr3/7jr3_refined.pdb | http://covid19.bioreproducibility.org/static/data/7jr3/7jr3_refined.mtz | 19.40% | Placed the coordinates in a standardized location in the unit cell using ACHESYM. | The coordinates are NOT in a standardized location in the unit cell. | 3C-like proteinase - UNP residues 3264-3569 | 64.8 | minimal | Yes | Yes | - | - | 90.9 | 48.2 | 65.4 | - | |||||
| 7jr4 | 1.55 Å | 2020-08-26 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 15.30% | 18.02% | 2.50% | - | XDS | 94.10% | 22.17 | LNLS SIRUS (MANACA) | PHENIX | 2020-08-11 | - | - | - | http://covid19.bioreproducibility.org/static/data/7jr4/7jr4_refined.pdb | http://covid19.bioreproducibility.org/static/data/7jr4/7jr4_refined.mtz | 18.70% | Placed the coordinates in a standardized location in the unit cell using ACHESYM. | The coordinates are NOT in a standardized location in the unit cell. | 3C-like proteinase - UNP residues 3264-3569 | 68.4 | minimal | Yes | Yes | - | - | 92.1 | 39.0 | 81.0 | - | |||||
| 7jrn | 2.48 Å | 2020-08-26 | X-ray | TTT | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 24.54% | 28.72% | 15.80% | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide | HKL-3000 | 86.20% | 5.4 | APS (19-BM) | REFMAC | 2020-08-12 | 34341772 | ZN | - | - | - | N/A | - | - | Papain-like proteinase | 32.2 | - | No | - | 11.0 | 48.4 | 81.1 | - | ||||||
| 7jtl | 2.04 Å | 2020-08-26 | X-ray | - | SARS-CoV-2 | ORF8 | No functional ligands | 21.99% | 26.40% | 10.09% | - | XDS | 95.06% | 14.92 | ALS (5.0.2) | PHENIX | 2020-08-18 | 33361333 | - | - | - | - | N/A | - | - | Nonstructural protein NS8 | 21.8 | - | No | - | 28.8 | 31.2 | 58.6 | - | |||||||
| 6xf5 | 3.45 Å | 2020-09-02 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-06-15 | 32742241 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6xf6 | 4.00 Å | 2020-09-02 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-06-15 | 32742241 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6zh9 | 3.31 Å | 2020-09-02 | X-ray | - | SARS-CoV-2 | Spike | No functional ligands | 26.09% | 30.52% | 9.10% | - | XDS | 100.00% | 0.018 | Diamond (I03) | REFMAC | 2020-06-21 | 32661423 | - | - | - | - | N/A | - | - | CR3022 heavy; CR3022 Light chain; Spike glycoprotein; Nanobody H11-H4 | 2.8 | - | No | - | 6.5 | 13.4 | 38.8 | - | |||||||
| 7c2q | 1.93 Å | 2020-09-02 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 22.62% | 26.50% | 11.80% | - | XDS | 99.90% | 15.4 | SSRF (BL17U1) | PHENIX | 2020-05-08 | 32880863 | - | - | http://covid19.bioreproducibility.org/static/data/7c2q/7c2q_rerefined.pdb | http://covid19.bioreproducibility.org/static/data/7c2q/7c2q_rerefined.mtz | 24.15% | Standardized with Achesym server; added NCS restraints; added over 250 water molecules; Fixed numerous sidechains and rotamers; added residues: we were able to trace GLN 299 and CYS 300 not mentioned in the FASTA file, but clearly visible and similar to other structures. | Structure practically unmodelled with many sidechains missing and few residures missing in both chains. | 3C-like proteinase | 23.4 | significant | Yes | Yes | - | 22.9 | 28.3 | 70.9 | - | ||||||
| 7c2y | 1.91 Å | 2020-09-02 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 22.01% | 26.17% | 4.90% | - | XDS | 99.90% | 13.9 | SSRF (BL17U1) | PHENIX | 2020-05-10 | - | - | - | http://covid19.bioreproducibility.org/static/data/7c2y/7c2y_rerefined.pdb | http://covid19.bioreproducibility.org/static/data/7c2y/7c2y_rerefined.mtz | 23.34% | Standardized with Achesym server; added NCS restraints; added over 250 water molecules; Fixed numerous sidechains and rotamers; added residues: we were able to trace GLN 299 and CYS 300 not mentioned in the FASTA file, but clearly visible and similar to other structures. | Structure practically unmodelled with many sidechains missing and few residures missing in both chains. | 3C-like proteinase | 43.3 | significant | Yes | Yes | - | - | 25.5 | 55.8 | 81.1 | - | |||||
| 7c6s | 1.60 Å | 2020-09-02 | X-ray | U5G | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.75% | 22.22% | N/A | boceprevir (bound form) | boceprevir (bound form) | HKL-2000 | 99.90% | 22.3 | SSRF (BL19U1) | PHENIX | 2020-05-22 | 32887884 | - | - | http://covid19.bioreproducibility.org/static/data/7c6s/7c6s_rerefined.pdb | http://covid19.bioreproducibility.org/static/data/7c6s/7c6s_rerefined.mtz | 16.57% | Standardized with Achesym server; added anisotropic refinement; deleted few water molecules, added over 220 water molecules; added residues: GLY 302, VAL 303, THR 304, PHE 305; modeled disorder for: LYS 12, GLU 47, SER 62, GLN 110, CYS 117, SER 267, GLU 288, ARG 298; fixed/changed rotamers for: GLN 19, THR 24, LEU 67, GLN 69, VAL 86, THR 190, ARG 217, THR 224, LYS 236, ASN 277; flipped or added sidechain for: ASN 53, ASN 84, GLN 119, ARG 222. | Many missing water molecules, unmodeled disorder, incorrect rotamers, missing residues. | 3C-like proteinase | 53.8 | significant | Yes | Yes | - | 62.9 | 42.2 | 77.4 | - | |||||
| 7c6u | 2.00 Å | 2020-09-02 | X-ray | K36 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 24.15% | 25.12% | N/A | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | HKL-2000 | 99.80% | 34.4 | SSRF (BL19U1) | PHENIX | 2020-05-22 | 32887884 | - | - | http://covid19.bioreproducibility.org/static/data/7c6u/7c6u_rerefined.pdb | http://covid19.bioreproducibility.org/static/data/7c6u/7c6u_rerefined.mtz | 23.40% | Achesym, re-refined in Refmac using identical TLS ranges as in the original file. Otherwise not corrected. | There is some indication of the low-occupancy alternate chirality of C21 in the link to Cys145, but we did not model it. | 3C-like proteinase | 23.7 | minimal | Yes | Yes | - | 34.0 | 29.1 | 59.5 | - | |||||
| 7c8b | 2.20 Å | 2020-09-02 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.70% | 23.00% | 6.60% | Z-VAD(OMe)-FMK | - | XDS | 99.80% | 21.4 | SSRF (BL19U1) | REFMAC | 2020-05-29 | - | - | - | http://covid19.bioreproducibility.org/static/data/7c8b/7c8b_refined.pdb | http://covid19.bioreproducibility.org/static/data/7c8b/7c8b_refined.mtz | 23.00% | Achesymed. Since the connection to Cys145 is very poor, it required strong restraints. | The connection to Cys145 is very poor. | 3C-like proteinase; Z-VAD(OMe)-FMK | 36.4 | minimal | Yes | Yes | - | - | 54.9 | 39.7 | 54.3 | - | ||||
| 7c8d | 3.00 Å | 2020-09-02 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | PHENIX | 2020-05-29 | 33020722 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike protein S1 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7cbt | 2.35 Å | 2020-09-02 | X-ray | K36 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.92% | 29.20% | N/A | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | HKL-3000 | 99.90% | 2.625 | ROTATING ANODE () | PHENIX | 2020-06-13 | 33691601 | - | - | http://covid19.bioreproducibility.org/static/data/7cbt/7cbt_rerefined.pdb | http://covid19.bioreproducibility.org/static/data/7cbt/7cbt_rerefined.mtz | 27.20% | Completely rebuilt chain between residues 210 and 290 in chain B. Added waters, including the “catalytic” water in chain A, flipped His41 bound to it (in both chains). Phe group of the inhibitor reoriented in chain B. Adjusted residues 55 and 222 in chain A, 47 and 137 in B, flipped peptide 194 in B. Applied TLS – for each whole each molecule. | Many issues with chain B. | Replicase polyprotein 1ab | 0.8 | significant | Yes | Yes | - | 9.3 | 14.6 | 18.7 | - | |||||
| 7cdz | 1.80 Å | 2020-09-02 | X-ray | - | SARS-CoV-2 | Nucleocapsid | No functional ligands | 19.73% | 22.66% | N/A | - | HKL-3000 | 98.40% | 32.724 | SSRF (BL19U1) | PHENIX | 2020-06-21 | 32914439 | - | - | - | - | N/A | - | - | Nucleoprotein - NTD | 21.0 | - | No | - | 58.6 | 14.3 | 43.9 | - | |||||||
| 7ce0 | 1.50 Å | 2020-09-02 | X-ray | - | SARS-CoV-2 | Nucleocapsid | No functional ligands | 17.67% | 19.03% | N/A | - | HKL-3000 | 91.70% | 30.265 | SSRF (BL19U1) | PHENIX | 2020-06-21 | 32914439 | - | - | - | - | N/A | - | - | Nucleoprotein - CTD | 56.7 | - | No | - | 87.6 | 42.2 | 58.3 | - | |||||||
| 7cjd | 2.50 Å | 2020-09-02 | X-ray | - | SARS-CoV-2 | NSP3: PLpro | No functional ligands | 23.31% | 28.19% | 18.30% | - | XDS | 99.20% | 9.53 | SSRF (BL17U) | PHENIX | 2020-07-10 | 32895623 | ZN | - | - | - | N/A | - | - | Replicase polyprotein 1ab | 2.3 | - | No | - | 13.3 | 18.0 | 24.0 | - | |||||||
| 7cjm | 3.20 Å | 2020-09-02 | X-ray | TTT | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 26.81% | 28.59% | 19.02% | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide | XDS | 99.05% | 12.92 | ROTATING ANODE () | PHENIX | 2020-07-11 | 33473130 | ZN | - | - | - | N/A | - | - | Non-structural protein 3 | 13.5 | - | No | - | 11.6 | 36.4 | 50.9 | - | ||||||
| 7cmd | 2.59 Å | 2020-09-02 | X-ray | TTT | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 24.37% | 29.80% | 31.80% | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide | XDS | 99.20% | 3.24 | SSRF (BL17U) | PHENIX | 2020-07-27 | 32895623 | ZN | - | - | - | N/A | - | - | Replicase polyprotein 1ab | 8.7 | - | No | - | 7.7 | 37.7 | 38.7 | - | ||||||
| 7ctt | 3.20 Å | 2020-09-02 | Cryo-EM | GE6 | SARS-CoV-2 | NSP7/NSP8/NSP12/RNA | Protein-protein complex | N/A | N/A | N/A | Primer-RNA (5'-R(P*UP*UP*CP*UP*CP*CP*UP*AP*AP*GP*AP*AP*GP*CP*UP*A)-3'); [[(2~{R},3~{S},4~{R},5~{R})-5-(3-aminocarbonyl-5-fluoranyl-2-oxidanylidene-pyrazin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate; Template-RNA (5'-R(P*AP*CP*UP*AP*GP*CP*UP*UP*CP*UP*UP*AP*GP*GP*AP*GP*AP*A)-3') | [[(2~{R},3~{S},4~{R},5~{R})-5-(3-aminocarbonyl-5-fluoranyl-2-oxidanylidene-pyrazin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | - | - | () | PHENIX | 2020-08-20 | 33521757 | MG; ZN | - | - | - | N/A | - | - | nsp12; nsp8; nsp7; Primer-RNA (5'-R(P*UP*UP*CP*UP*CP*CP*UP*AP*AP*GP*AP*AP*GP*CP*UP*A)-3'); Template-RNA (5'-R(P*AP*CP*UP*AP*GP*CP*UP*UP*CP*UP*UP*AP*GP*GP*AP*GP*AP*A)-3') | N/A | - | No | - | N/A | N/A | N/A | Primer-RNA (5'-R(P*UP*UP*CP*UP*CP*CP*UP*AP*AP*GP*AP*AP*GP*CP*UP*A)-3'); Template-RNA (5'-R(P*AP*CP*UP*AP*GP*CP*UP*UP*CP*UP*UP*AP*GP*GP*AP*GP*AP*A)-3') | - | ||||||
| 7jme | 1.55 Å | 2020-09-02 | X-ray | CMP | SARS-CoV-2 | NSP3: Macro | Functional ligand | 15.68% | 18.21% | 7.64% | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE | HKL-2000 | 95.05% | 10.91 | APS (21-ID-F) | PHENIX | 2020-07-31 | 32981460 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 80.2 | - | No | - | 91.3 | 55.3 | 96.7 | - | ||||||
| 7jun | 2.30 Å | 2020-09-02 | X-ray/N-diffr. | DOD | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 18.50% | 22.00% | 12.90% | water | Mantid; CrysalisPro | 97.90% | 13.0 | ROTATING ANODE, SPALLATION SOURCE () | nCNS | 2020-08-20 | 33060199 | - | - | - | - | N/A | - | - | 3C-like proteinase | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7ju7 | 1.60 Å | 2020-09-02 | X-ray | G65 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.86% | 19.18% | 8.40% | Masitinib | Masitinib | HKL-3000 | 98.50% | 26.3 | APS (19-ID) | PHENIX | 2020-08-20 | 34285133 | - | - | http://covid19.bioreproducibility.org/static/data/7ju7/7ju7_rerefinement.pdb | http://covid19.bioreproducibility.org/static/data/7ju7/7ju7_rerefinement.mtz | 17.13% | Standardized unit cell placement by running Achesym; applied anisotropic refinement; added few alternative conformations to several sidechains; added 91 water molecules that appeared on electron density map when anisotropic refinement was applied. | Structure looks generally good; few missing alternative conformations; the electron density on some parts of the ligand is weak but the placement of the ligand is justified. | 3C-like proteinase | 72.4 | minimal | Yes | Yes | - | 86.8 | 42.5 | 92.1 | - | |||||
| 7jyc | 1.79 Å | 2020-09-09 | X-ray | NNA, NA | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.50% | 21.25% | 7.90% | SODIUM ION; (1R,2S,5S)-3-[N-({1-[(tert-butylsulfonyl)methyl]cyclohexyl}carbamoyl)-3-methyl-L-valyl]-N-{(1S)-1-[(1R)-2-(cyclopropyla mino)-1-hydroxy-2-oxoethyl]pentyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-3-[N-({1-[(tert-butylsulfonyl)methyl]cyclohexyl}carbamoyl)-3-methyl-L-valyl]-N-{(1S)-1-[(1R)-2-(cyclopropyla mino)-1-hydroxy-2-oxoethyl]pentyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide; SODIUM ION | XDS | 98.00% | 10.5 | NSLS-II (17-ID-2) | REFMAC | 2020-08-19 | 35842458 | - | - | http://covid19.bioreproducibility.org/static/data/7jyc/7jyc_rerefinement.pdb | http://covid19.bioreproducibility.org/static/data/7jyc/7jyc_rerefinement.mtz | 21.44% | Minor changes; few rotamers changed; few sidechains flipped, added ~ 50 water molecules, changed occupancy form 0.1 to 1.0 for residues 302-306. | Overall good structure, with only some wierd 10% occupancy at the end of the main chain. | 3C-like proteinase | 52.5 | minimal | Yes | Yes | - | 72.0 | 52.3 | 55.6 | - | |||||
| 7a5r | 3.70 Å | 2020-09-09 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | 2020-08-30 | 33087721 | - | - | - | - | N/A | - | - | CR3022 Fab Heavy Chain; CR3022 Fab Light Chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7a5s | 3.90 Å | 2020-09-16 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | 2020-08-21 | 33087721 | - | - | - | - | N/A | - | - | CR3022 Fab Heavy Chain; CR3022 Fab Light Chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7a91 | 3.60 Å | 2020-09-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-08-21 | 32942285 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7a93 | 5.90 Å | 2020-09-16 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2020-09-01 | 32942285 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7a94 | 3.90 Å | 2020-09-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-09-01 | 32942285 | ZN | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7a95 | 4.30 Å | 2020-09-16 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2020-09-01 | 32942285 | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7a96 | 4.80 Å | 2020-09-16 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2020-09-01 | 32942285 | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7a97 | 4.40 Å | 2020-09-16 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2020-09-01 | 32942285 | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7ch4 | 3.15 Å | 2020-09-16 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 22.53% | 27.26% | N/A | - | HKL-2000 | 100.00% | 7.78 | SSRF (BL19U1) | PHENIX | 2020-09-01 | 32970990 | - | - | - | - | N/A | - | - | BD-604 Fab H; BD-604 Fab L; Spike glycoprotein | 33.9 | - | No | - | 18.1 | 69.3 | 56.4 | - | |||||||
| 7ch5 | 2.70 Å | 2020-09-16 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 22.69% | 25.98% | N/A | - | HKL-2000 | 100.00% | 13.78 | SSRF (BL17U1) | PHENIX | 2020-07-05 | 32970990 | - | - | - | - | N/A | - | - | BD-629 Fab H; BD-629 Fab L; Spike glycoprotein | 28.9 | - | No | - | 26.8 | 48.6 | 58.1 | - | |||||||
| 7chb | 2.40 Å | 2020-09-16 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 19.24% | 22.56% | N/A | - | HKL-2000 | 99.62% | 15.2 | SSRF (BL17U1) | PHENIX | 2020-07-05 | 32970990 | - | - | - | - | N/A | - | - | BD-236 Fab heavy chain; BD-236 Fab light chain; SARS-coV-2 Receptor Binding Domain | 48.1 | - | No | - | 59.7 | 47.4 | 64.4 | - | |||||||
| 7chc | 2.71 Å | 2020-09-16 | X-ray | - | SARS-CoV-2 | Spike | Protein-protein complex | 21.21% | 28.58% | N/A | - | HKL-2000 | 99.90% | 12.49 | SSRF (BL17U1) | PHENIX | 2020-07-05 | 32970990 | - | - | - | - | N/A | - | - | BD-629 Fab H; IGK@ protein; Spike glycoprotein; BD-368-2 Fab H; BD-368-2 Fab L | 25.4 | - | No | - | 11.6 | 69.6 | 45.2 | - | |||||||
| 7che | 3.42 Å | 2020-09-16 | X-ray | - | SARS-CoV-2 | Spike | Protein-protein complex | 22.47% | 23.88% | N/A | - | HKL-2000 | 94.54% | 11.55 | SSRF (BL17U1) | PHENIX | 2020-07-05 | 32970990 | - | - | - | - | N/A | - | - | BD-236 Fab heavy chain; BD-236 Fab light chain; SARS-coV-2 Receptor Binding Domain; BD-368-2 Fab heavy chain; BD-368-2 Fab light chain | 20.2 | - | No | - | 46.1 | 60.6 | 8.2 | - | |||||||
| 7chf | 2.67 Å | 2020-09-16 | X-ray | - | SARS-CoV-2 | Spike | Protein-protein complex | 21.96% | 26.98% | N/A | - | HKL-2000 | 95.96% | 16.2 | SSRF (BL17U1) | PHENIX | 2020-07-05 | 32970990 | - | - | - | - | N/A | - | - | BD-604 Fab heavy chain; BD-604 Fab light chain; SARS-coV-2 Receptor Binding Domain; BD-368-2 Fab heavy chain; BD-368-2 Fab light chain | 12.5 | - | No | - | 19.8 | 50.7 | 25.4 | - | |||||||
| 7chh | 3.49 Å | 2020-09-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-07-05 | 32970990 | - | - | - | - | N/A | - | - | Spike glycoprotein; BD-368-2 Fab heavy chain; BD-368-2 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7cx9 | 1.73 Å | 2020-09-16 | X-ray | GKF | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.01% | 20.92% | 5.70% | 3-iodanyl-1~{H}-indazole-7-carbaldehyde | 3-iodanyl-1~{H}-indazole-7-carbaldehyde | XDS | 97.20% | 21.6 | SSRF (BL19U1) | REFMAC | 2020-07-05 | - | - | - | http://covid19.bioreproducibility.org/static/data/7cx9/7cx9_refined.pdb | http://covid19.bioreproducibility.org/static/data/7cx9/7cx9_refined.mtz | 18.83% | Achesymed. Added missing residues 45 and 46 and reordered 301 at the C terminus. Removed double conformation of residues 110, 142, 155, 198, 225, 226, 245, 270. Deleted 16 waters, added 2. Refined with 6 TLS groups. The occupancy of the I atom of the inhibitor GKF was originally set at 0.51, and changed that to 1.0, but there is very large negative density. Possible interpretation is radiation damage. | Missing residues, unnecessary double conformation. | 3C protein | 37.6 | minimal | Yes | Yes | - | - | 74.7 | 30.2 | 46.3 | - | ||||
| 7jvz | 2.50 Å | 2020-09-16 | XFEL | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 17.23% | 21.72% | N/A | - | CrystFEL | 100.00% | 5.0 | FREE ELECTRON LASER () | REFMAC | 2020-09-01 | - | - | - | http://covid19.bioreproducibility.org/static/data/7jvz/7jvz_refined.pdb | http://covid19.bioreproducibility.org/static/data/7jvz/7jvz_refined.mtz | 25.20% | Placed the coordinates in a standardized location in the unit cell using ACHESYM. | The coordinates are NOT in a standardized location in the unit cell. | 3C-like proteinase | N/A | minimal | Yes | Yes | - | - | N/A | N/A | N/A | - | |||||
| 7jyy | 2.05 Å | 2020-09-16 | X-ray | SAM, NA, FMT | SARS-CoV-2 | NSP10/NSP16/RNA | Functional ligand | 16.58% | 18.46% | 8.30% | FORMIC ACID; SODIUM ION; S-ADENOSYLMETHIONINE; RNA (5'-D(*(M7G))-R(P*AP*UP*UP*A)-3') | S-ADENOSYLMETHIONINE; SODIUM ION; FORMIC ACID | HKL-3000 | 100.00% | 24.1 | APS (21-ID-F) | REFMAC | 2020-08-24 | 34131072 | NA; MG; ZN | - | http://covid19.bioreproducibility.org/static/data/7jyy/7jyy_rerefinement2.pdb | http://covid19.bioreproducibility.org/static/data/7jyy/7jyy_rerefinement2.mtz | 17.85% | ACHESYM, NCS, TLS (based the ranges on the deposited pdb file). Added external restraints from RetraintLib. Large unmodeled electon density next to TYR 7020 A, probably a nucleotide. Nucleotides in chains E and F should have partial occupancy, since there many negative electron density peaks and B-factors seems to be high. The Rfree improvement is achieved mainly by relaxing restraints, with the increase of RMS deviations for bonds and angles. There were inconsistencies between 7jyy and 7jz0 after achesym, so the grouping was changed to (A,B,E;C,D,F) so the chains are in the same place. | Overly restrained structure, requires ACHESYM, NCS, and TLS. | 2'-O-methyltransferase; Non-structural protein 10; RNA (5'-D(*(M7G))-R(P*AP*UP*UP*A)-3') | 59.7 | minimal | Yes | Yes | 90.4 | 13.8 | 89.8 | RNA (5'-D(*(M7G))-R(P*AP*UP*UP*A)-3') | - | |||||
| 7jz0 | 2.15 Å | 2020-09-16 | X-ray | SAH, NA, FMT | SARS-CoV-2 | NSP10/NSP16/RNA | Functional ligand | 17.33% | 19.81% | 9.00% | FORMIC ACID; RNA (5'-D(*(M7G))-R(P*(A2M)P*UP*U)-3'); SODIUM ION; S-ADENOSYL-L-HOMOCYSTEINE | S-ADENOSYL-L-HOMOCYSTEINE; SODIUM ION; FORMIC ACID | HKL-3000 | 100.00% | 26.6 | APS (21-ID-F) | REFMAC | 2020-09-01 | - | ZN; NA | - | http://covid19.bioreproducibility.org/static/data/7jz0/7jz0_rerefined.pdb | http://covid19.bioreproducibility.org/static/data/7jz0/7jz0_rerefined.mtz | 19.66% | ACHESYM , NCS, TLS, and external restraints from RetraintLib. Nucleotides in chains E and F should have partial occupancy, since there are many negative electron density peaks and B-factors seem to be high. The Rfree improvement was achieved mainly by relaxing restraints, with the increase of RMS deviations for bonds and angles. No manual changes. | Overly restrained structure, requires ACHESYM, NCS, and TLS. | 2'-O-methyltransferase; Non-structural protein 10; RNA (5'-D(*(M7G))-R(P*(A2M)P*UP*U)-3') | 57.5 | minimal | Yes | Yes | - | 82.9 | 12.6 | 94.0 | RNA (5'-D(*(M7G))-R(P*(A2M)P*UP*U)-3') | - | ||||
| 6zxn | 2.93 Å | 2020-09-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-09-01 | 32887876 | - | - | - | - | N/A | - | - | Spike glycoprotein; Nanobody Ty1 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7a98 | 5.40 Å | 2020-09-23 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2020-07-30 | 32942285 | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7aap | 2.50 Å | 2020-09-23 | Cryo-EM | POP, GE6 | SARS-CoV-2 | NSP7/NSP8/NSP12/RNA | Protein-protein complex | N/A | N/A | N/A | RNA (5'-R(P*UP*UP*AP*AP*GP*UP*UP*AP*U)-3'); [[(2~{R},3~{S},4~{R},5~{R})-5-(3-aminocarbonyl-5-fluoranyl-2-oxidanylidene-pyrazin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate; PYROPHOSPHATE 2-; RNA (5'-R(P*UP*UP*CP*AP*UP*AP*AP*CP*UP*UP*AP*A)-3') | PYROPHOSPHATE 2-; [[(2~{R},3~{S},4~{R},5~{R})-5-(3-aminocarbonyl-5-fluoranyl-2-oxidanylidene-pyrazin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | - | - | () | PHENIX; phenix.real_space_refine | 2020-09-01 | 33526596 | ZN; MG | - | - | - | N/A | - | - | Non-structural protein 12; Non-structural protein 8; Non-structural protein 7; RNA (5'-R(P*UP*UP*AP*AP*GP*UP*UP*AP*U)-3'); RNA (5'-R(P*UP*UP*CP*AP*UP*AP*AP*CP*UP*UP*AP*A)-3') | N/A | - | No | - | N/A | N/A | N/A | RNA (5'-R(P*UP*UP*CP*AP*UP*AP*AP*CP*UP*UP*AP*A)-3'); RNA (5'-R(P*UP*UP*AP*AP*GP*UP*UP*AP*U)-3') | - | ||||||
| 7cai | 3.49 Å | 2020-09-23 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-09-04 | 32703908 | - | - | - | - | N/A | - | - | Spike glycoprotein - UNP residues 1-1208; Light chain of H014 Fab; Heavy chain of H014 Fab | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7cak | 3.58 Å | 2020-09-23 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-06-08 | 32703908 | - | - | - | - | N/A | - | - | Spike glycoprotein - UNP residues 1-1208; Light chain of H014 Fab; Heavy chain of H014 Fab | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7d1o | 1.78 Å | 2020-09-23 | X-ray | NNA | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 22.28% | 24.85% | N/A | (1R,2S,5S)-3-[N-({1-[(tert-butylsulfonyl)methyl]cyclohexyl}carbamoyl)-3-methyl-L-valyl]-N-{(1S)-1-[(1R)-2-(cyclopropyla mino)-1-hydroxy-2-oxoethyl]pentyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-3-[N-({1-[(tert-butylsulfonyl)methyl]cyclohexyl}carbamoyl)-3-methyl-L-valyl]-N-{(1S)-1-[(1R)-2-(cyclopropyla mino)-1-hydroxy-2-oxoethyl]pentyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | HKL-2000 | 99.70% | 16.8 | SSRF (BL17U1) | PHENIX | 2020-06-08 | 33542181 | - | - | - | - | N/A | - | - | 3C-like proteinase | 30.9 | - | No | - | 36.7 | 32.6 | 68.5 | - | ||||||
| 7jkv | 1.25 Å | 2020-09-23 | X-ray | V7G, 1PE | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 14.46% | 17.69% | 5.87% | PENTAETHYLENE GLYCOL; N-[(2S)-1-({(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | N-[(2S)-1-({(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide; PENTAETHYLENE GLYCOL | xia2 | 95.08% | 10.52 | SPring-8 (BL24XU) | REFMAC | 2020-09-15 | 33510133 | - | - | - | - | N/A | - | - | 3C-like proteinase | 70.1 | - | No | - | 93.3 | 64.1 | 58.6 | - | ||||||
| 7jx6 | 1.61 Å | 2020-09-23 | X-ray | - | SARS-CoV-2 | ORF8 | No functional ligands | 20.23% | 23.31% | 5.65% | SODIUM ION | SODIUM ION | XDS | 99.76% | 27.09 | ALS (4.2.2) | PHENIX | 2020-07-29 | - | - | - | http://covid19.bioreproducibility.org/static/data/7jx6/7jx6_rerefinement.pdb | http://covid19.bioreproducibility.org/static/data/7jx6/7jx6_rerefinement.mtz | 21.59% | Standardized unit cell placement by running Achesym; applied anisotropic refinement; added waters, added disorder to ASP 34A, ASP 34B, ILE 39 B, GLU 106 A; added OXT to ILE 121 A and ILE 121 ; changed rotamers: ILE 47 B, ARG 52 B, LYS 53 B, ILE 58 A, VAL 62 A, LYS 94 A. | Some missing disorder, several rotamers to fix and missing residues. Residues 63-78 in chain B are missing, and it was possible to trace some parts of the missing model; however, the B factors were very high and did not improve the R factor enough to model that fragment. It seems there is a bigger electron density next to GLN 18 A, but we were not able to find a good solution and keep waters in the place. | ORF8 protein | 49.2 | minimal | Yes | Yes | - | - | 51.9 | 53.9 | 67.7 | - | ||||
| 7jzl | 2.70 Å | 2020-09-23 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-08-26 | 32907861 | - | - | - | - | N/A | - | - | Spike glycoprotein; LCB1 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7jzm | 3.50 Å | 2020-09-23 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2020-09-02 | 32907861 | - | - | - | - | N/A | - | - | LCB3; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7jzn | 3.10 Å | 2020-09-23 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-09-02 | 32907861 | - | - | - | - | N/A | - | - | Spike glycoprotein; LCB3 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7jzu | 3.10 Å | 2020-09-23 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2020-09-02 | 32907861 | - | - | - | - | N/A | - | - | LCB1; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7k1l | 2.25 Å | 2020-09-23 | X-ray | UVC | SARS-CoV-2 | NSP15 | Functional ligand | 16.66% | 19.15% | 19.30% | URIDINE-2',3'-VANADATE | URIDINE-2',3'-VANADATE | HKL-3000 | 98.60% | 10.8 | APS (19-ID) | PHENIX | 2020-09-02 | 33564093 | - | - | http://covid19.bioreproducibility.org/static/data/7k1l/7k1l_achesym-coot-73-hoh_pdbin.pdb | http://covid19.bioreproducibility.org/static/data/7k1l/7k1l_refined.mtz | 16.79% | Achesymed. Added SO4, ACT, replaced SO4 with TRS, added 2 EDO and 74 HOH. Changes several rotamers. | ASN 0 -> SER 2 in A and B should be disordered. HOH 502 A (next to UVC 401 A) has a bond to V , not sure if it is the right metal coordination. Next to UVC 401 B there is unmodeled electron density. In chain UVC 401 A there is ACY in that position. | Uridylate-specific endoribonuclease | 85.2 | moderate | Yes | Yes | - | 87.0 | 93.8 | 85.5 | - | |||||
| 7k1o | 2.40 Å | 2020-09-23 | X-ray | VQV | SARS-CoV-2 | NSP15 | Functional ligand | 20.68% | 24.25% | 14.40% | 1-(3,5-di-O-phosphono-alpha-L-xylofuranosyl)pyrimidine-2,4(1H,3H)-dione | 1-(3,5-di-O-phosphono-alpha-L-xylofuranosyl)pyrimidine-2,4(1H,3H)-dione | HKL-3000 | 96.80% | 15.0 | APS (19-ID) | PHENIX | 2020-09-07 | - | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 59.3 | - | No | - | - | 42.5 | 65.5 | 85.0 | - | |||||
| 7k3t | 1.20 Å | 2020-09-23 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 17.10% | 18.66% | 9.40% | - | XDS | 97.80% | 11.6 | NSLS-II (17-ID-2) | REFMAC | 2020-09-08 | 35842458 | - | - | http://covid19.bioreproducibility.org/static/data/7k3t/7k3t_refined.pdb | http://covid19.bioreproducibility.org/static/data/7k3t/7k3t_refined.mtz | 16.30% | Re-indexed to C2 space group, ACHESYMED, added several water molecules, employed full anysotropic refinement, modified residues 97, 102, 269, 189, added PEG | I2 space group. Too high R-free for 1.2A resulution | 3C-like proteinase | 71.4 | significant | Yes | Yes | - | 95.7 | 40.0 | 83.2 | http://covid19.bioreproducibility.org/static/data/7k3t/7k3t_refined_report.pdf | ||||||
| 7k40 | 1.35 Å | 2020-09-23 | X-ray | U5G | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.98% | 19.15% | 4.50% | boceprevir (bound form) | boceprevir (bound form) | XDS | 99.20% | 16.4 | NSLS-II (17-ID-2) | REFMAC | 2020-09-13 | 35842458 | - | - | http://covid19.bioreproducibility.org/static/data/7k40/7k40_refined.pdb | http://covid19.bioreproducibility.org/static/data/7k40/7k40_refined.mtz | 18.30% | Standardized unit cell placement by running Achesym; removed very model at very weak density after Cys300; change DMSO 403-406 to water molecules, added additional water moecules. | Improper cif file used for inhibitor | 3C-like proteinase - UNP residues 3264-3569 | 51.3 | moderate | Yes | Yes | - | 87.0 | 29.5 | 61.1 | - | |||||
| 5rl6 | 1.92 Å | 2020-09-23 | X-ray | PO4, LJA | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 17.82% | 24.95% | 6.70% | PHOSPHATE ION; N-[3-(carbamoylamino)phenyl]acetamide | PHOSPHATE ION; N-[3-(carbamoylamino)phenyl]acetamide | XDS | 96.80% | 8.2 | Diamond (I04-1) | BUSTER | 2020-09-14 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 13.4 | - | No | - | 35.7 | 14.3 | 48.5 | - | ||||||
| 5rl7 | 1.89 Å | 2020-09-30 | X-ray | VVD, PO4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 22.68% | 26.90% | 6.50% | 5-(acetylamino)-2-fluorobenzoic acid; PHOSPHATE ION | 5-(acetylamino)-2-fluorobenzoic acid; PHOSPHATE ION | XDS | 96.50% | 8.6 | Diamond (I04-1) | REFMAC | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 6.2 | - | No | - | 20.1 | 15.2 | 39.4 | - | ||||||
| 5rl8 | 2.21 Å | 2020-09-30 | X-ray | VVG, PO4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 20.94% | 26.95% | 12.20% | N-(2-fluorophenyl)ethanesulfonamide; PHOSPHATE ION | N-(2-fluorophenyl)ethanesulfonamide; PHOSPHATE ION | XDS | 97.50% | 5.7 | Diamond (I04-1) | REFMAC | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 12.1 | - | No | - | 19.9 | 39.3 | 35.7 | - | ||||||
| 5rl9 | 1.79 Å | 2020-09-30 | X-ray | PO4, UR7 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 17.70% | 23.14% | 7.10% | PHOSPHATE ION; 1-(3-fluoro-4-methylphenyl)methanesulfonamide | PHOSPHATE ION; 1-(3-fluoro-4-methylphenyl)methanesulfonamide | XDS | 96.30% | 7.1 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 27.2 | - | No | - | 53.6 | 16.8 | 59.5 | - | ||||||
| 5rlb | 1.98 Å | 2020-09-30 | X-ray | VVJ, PO4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 21.40% | 26.19% | 9.60% | N-cycloheptyl-N-methylmethanesulfonamide; PHOSPHATE ION | N-cycloheptyl-N-methylmethanesulfonamide; PHOSPHATE ION | XDS | 96.30% | 6.7 | Diamond (I04-1) | REFMAC | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 12.3 | - | No | - | 25.3 | 36.1 | 34.0 | - | ||||||
| 5rlc | 1.92 Å | 2020-09-30 | X-ray | PO4, VVM | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 20.60% | 25.22% | 6.60% | PHOSPHATE ION; 4-amino-N-phenylbenzene-1-sulfonamide | PHOSPHATE ION; 4-amino-N-phenylbenzene-1-sulfonamide | XDS | 96.80% | 8.3 | Diamond (I04-1) | REFMAC | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 19.0 | - | No | - | 33.2 | 33.4 | 45.5 | - | ||||||
| 5rld | 2.23 Å | 2020-09-30 | X-ray | PO4, VVY | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 16.62% | 26.96% | 11.50% | PHOSPHATE ION; 2-phenoxy-1-(pyrrolidin-1-yl)ethan-1-one | PHOSPHATE ION; 2-phenoxy-1-(pyrrolidin-1-yl)ethan-1-one | XDS | 96.80% | 5.7 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 10.6 | - | No | - | 19.8 | 32.8 | 37.5 | - | ||||||
| 5rle | 2.27 Å | 2020-09-30 | X-ray | PO4, VVP | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 15.14% | 26.16% | 8.50% | PHOSPHATE ION; 4-methoxy-1H-indole | PHOSPHATE ION; 4-methoxy-1H-indole | XDS | 96.30% | 7.0 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 8.4 | - | No | - | 25.5 | 22.2 | 35.3 | - | ||||||
| 5rlf | 2.23 Å | 2020-09-30 | X-ray | NY7, PO4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 16.07% | 25.39% | 7.80% | N-(2-methoxy-5-methylphenyl)glycinamide; PHOSPHATE ION | N-(2-methoxy-5-methylphenyl)glycinamide; PHOSPHATE ION | XDS | 96.80% | 8.1 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 14.1 | - | No | - | 31.9 | 23.8 | 44.7 | - | ||||||
| 5rlg | 1.96 Å | 2020-09-30 | X-ray | VW1, PO4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 24.58% | 28.59% | 8.90% | (2S)-2-(4-cyanophenoxy)propanamide; PHOSPHATE ION | (2S)-2-(4-cyanophenoxy)propanamide; PHOSPHATE ION | XDS | 96.40% | 6.8 | Diamond (I04-1) | REFMAC | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 4.4 | - | No | - | 11.6 | 13.7 | 42.1 | - | ||||||
| 5rlh | 2.38 Å | 2020-09-30 | X-ray | PO4, K2P | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 15.61% | 25.08% | 16.80% | PHOSPHATE ION; 2-(trifluoromethoxy)benzoic acid | PHOSPHATE ION; 2-(trifluoromethoxy)benzoic acid | XDS | 97.20% | 5.5 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 14.8 | - | No | - | 34.6 | 28.6 | 38.9 | - | ||||||
| 5rli | 2.26 Å | 2020-09-30 | X-ray | JFM, PO4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 19.67% | 26.81% | 13.20% | N-(2-phenylethyl)methanesulfonamide; PHOSPHATE ION | N-(2-phenylethyl)methanesulfonamide; PHOSPHATE ION | XDS | 97.10% | 6.2 | Diamond (I04-1) | REFMAC | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 12.6 | - | No | - | 20.7 | 37.3 | 38.2 | - | ||||||
| 5rlj | 1.88 Å | 2020-09-30 | X-ray | PO4, VW4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 17.30% | 22.80% | 7.10% | PHOSPHATE ION; (2S)-2-phenylpropane-1-sulfonamide | PHOSPHATE ION; (2S)-2-phenylpropane-1-sulfonamide | XDS | 96.40% | 8.5 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 30.5 | - | No | - | 57.0 | 22.9 | 57.1 | - | ||||||
| 5rlk | 1.96 Å | 2020-09-30 | X-ray | PO4, NYV | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 17.25% | 23.97% | 7.40% | PHOSPHATE ION; 1-(propan-2-yl)-1H-imidazole-4-sulfonamide | PHOSPHATE ION; 1-(propan-2-yl)-1H-imidazole-4-sulfonamide | XDS | 96.60% | 7.8 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 23.5 | - | No | - | 45.2 | 26.1 | 51.0 | - | ||||||
| 5rll | 2.08 Å | 2020-09-30 | X-ray | PO4, H04 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 21.54% | 26.73% | 10.50% | PHOSPHATE ION; 1-(2-ethoxyphenyl)piperazine | PHOSPHATE ION; 1-(2-ethoxyphenyl)piperazine | XDS | 96.70% | 5.9 | Diamond (I04-1) | REFMAC | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 11.9 | - | No | - | 21.3 | 32.4 | 40.6 | - | ||||||
| 5rlm | 1.86 Å | 2020-09-30 | X-ray | PO4, VW7 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 17.28% | 23.67% | 7.20% | PHOSPHATE ION; N-(8-methyl-1,2,3,4-tetrahydroquinolin-5-yl)acetamide | PHOSPHATE ION; N-(8-methyl-1,2,3,4-tetrahydroquinolin-5-yl)acetamide | XDS | 96.10% | 7.3 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 23.7 | - | No | - | 48.3 | 22.2 | 52.2 | - | ||||||
| 5rln | 2.15 Å | 2020-09-30 | X-ray | NZG, PO4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 17.41% | 25.84% | 11.40% | 3-(acetylamino)-4-fluorobenzoic acid; PHOSPHATE ION | 3-(acetylamino)-4-fluorobenzoic acid; PHOSPHATE ION | XDS | 97.00% | 6.7 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 10.4 | - | No | - | 27.9 | 18.7 | 43.2 | - | ||||||
| 5rlo | 2.10 Å | 2020-09-30 | X-ray | PO4, UQS | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 16.71% | 24.28% | 10.20% | PHOSPHATE ION; N-[(2-fluorophenyl)methyl]-1H-pyrazol-4-amine | PHOSPHATE ION; N-[(2-fluorophenyl)methyl]-1H-pyrazol-4-amine | XDS | 97.10% | 5.9 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 15.9 | - | No | - | 42.3 | 20.2 | 42.4 | - | ||||||
| 5rlp | 2.56 Å | 2020-09-30 | X-ray | PO4, VWA | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 14.12% | 25.27% | 14.80% | PHOSPHATE ION; (1S)-1-(4-fluorophenyl)-N-methylethan-1-amine | PHOSPHATE ION; (1S)-1-(4-fluorophenyl)-N-methylethan-1-amine | XDS | 97.40% | 6.4 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 17.6 | - | No | - | 32.9 | 36.1 | 40.0 | - | ||||||
| 5rlq | 2.23 Å | 2020-09-30 | X-ray | UVA, PO4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 15.45% | 25.39% | 8.30% | N-methyl-2-(methylsulfonyl)aniline; PHOSPHATE ION | N-methyl-2-(methylsulfonyl)aniline; PHOSPHATE ION | XDS | 96.40% | 7.3 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 18.6 | - | No | - | 31.9 | 32.4 | 47.2 | - | ||||||
| 5rlr | 2.32 Å | 2020-09-30 | X-ray | PO4, VWD | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 20.88% | 28.26% | 12.10% | PHOSPHATE ION; (1R)-2-(methylsulfonyl)-1-phenylethan-1-ol | PHOSPHATE ION; (1R)-2-(methylsulfonyl)-1-phenylethan-1-ol | XDS | 96.40% | 7.3 | Diamond (I04-1) | REFMAC | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 8.3 | - | No | - | 12.9 | 36.7 | 33.1 | - | ||||||
| 5rls | 2.28 Å | 2020-09-30 | X-ray | VWG, PO4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 15.64% | 25.41% | 12.90% | N-hydroxyquinoline-2-carboxamide; PHOSPHATE ION | N-hydroxyquinoline-2-carboxamide; PHOSPHATE ION | XDS | 96.90% | 5.5 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 14.9 | - | No | - | 31.5 | 34.4 | 36.4 | - | ||||||
| 5rlt | 2.43 Å | 2020-09-30 | X-ray | PO4, UVJ | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 15.08% | 26.35% | 14.70% | PHOSPHATE ION; 3-(2-methyl-1H-benzimidazol-1-yl)propanamide | PHOSPHATE ION; 3-(2-methyl-1H-benzimidazol-1-yl)propanamide | XDS | 97.60% | 5.6 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 17.2 | - | No | - | 24.1 | 40.7 | 43.1 | - | ||||||
| 5rlu | 2.35 Å | 2020-09-30 | X-ray | JG4, PO4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 15.97% | 27.31% | 12.00% | 2-(thiophen-2-yl)-1H-imidazole; PHOSPHATE ION | 2-(thiophen-2-yl)-1H-imidazole; PHOSPHATE ION | XDS | 97.30% | 5.7 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 3.8 | - | No | - | 17.7 | 15.3 | 31.4 | - | ||||||
| 5rlv | 2.21 Å | 2020-09-30 | X-ray | VWJ, PO4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 20.10% | 25.96% | 10.60% | N-(propan-2-yl)-1H-benzimidazol-2-amine; PHOSPHATE ION | N-(propan-2-yl)-1H-benzimidazol-2-amine; PHOSPHATE ION | XDS | 96.70% | 7.6 | Diamond (I04-1) | REFMAC | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 14.0 | - | No | - | 26.9 | 37.3 | 35.9 | - | ||||||
| 5rlw | 1.97 Å | 2020-09-30 | X-ray | S9S, PO4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 17.11% | 23.72% | 10.10% | ~{N}-[2-(4-fluorophenyl)ethyl]methanesulfonamide; PHOSPHATE ION | ~{N}-[2-(4-fluorophenyl)ethyl]methanesulfonamide; PHOSPHATE ION | XDS | 96.30% | 6.7 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 21.7 | - | No | - | 47.6 | 23.2 | 47.3 | - | ||||||
| 5rly | 2.43 Å | 2020-09-30 | X-ray | K34, PO4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 15.44% | 26.05% | 13.60% | 5-(1,3-thiazol-2-yl)-1H-1,2,4-triazole; PHOSPHATE ION | 5-(1,3-thiazol-2-yl)-1H-1,2,4-triazole; PHOSPHATE ION | XDS | 96.90% | 6.4 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 17.8 | - | No | - | 26.2 | 47.7 | 35.7 | - | ||||||
| 5rlz | 1.97 Å | 2020-09-30 | X-ray | VWM, PO4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 22.15% | 27.34% | 8.60% | (3R)-1-acetyl-3-hydroxypiperidine-3-carboxylic acid; PHOSPHATE ION | (3R)-1-acetyl-3-hydroxypiperidine-3-carboxylic acid; PHOSPHATE ION | XDS | 96.40% | 6.4 | Diamond (I04-1) | REFMAC | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 8.2 | - | No | - | 17.6 | 26.4 | 38.5 | - | ||||||
| 5rm0 | 1.91 Å | 2020-09-30 | X-ray | PO4, S7G | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 16.78% | 23.85% | 7.60% | PHOSPHATE ION; ~{N}-[(3~{R})-1,2,3,4-tetrahydroquinolin-3-yl]ethanamide | PHOSPHATE ION; ~{N}-[(3~{R})-1,2,3,4-tetrahydroquinolin-3-yl]ethanamide | XDS | 96.00% | 6.9 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 21.1 | - | No | - | 46.4 | 26.4 | 44.3 | - | ||||||
| 5rm1 | 1.90 Å | 2020-09-30 | X-ray | PO4, RY4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 17.56% | 23.40% | 7.50% | PHOSPHATE ION; N-[4-(aminomethyl)phenyl]methanesulfonamide | PHOSPHATE ION; N-[4-(aminomethyl)phenyl]methanesulfonamide | XDS | 95.80% | 7.6 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 33.1 | - | No | - | 50.9 | 37.9 | 53.4 | - | ||||||
| 5rm2 | 1.82 Å | 2020-09-30 | X-ray | PO4, UXG | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 23.05% | 27.29% | 7.20% | PHOSPHATE ION; 1-(diphenylmethyl)azetidin-3-ol | PHOSPHATE ION; 1-(diphenylmethyl)azetidin-3-ol | XDS | 95.50% | 7.2 | Diamond (I04-1) | REFMAC | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 9.6 | - | No | - | 18.0 | 19.4 | 49.8 | - | ||||||
| 5rm3 | 2.09 Å | 2020-09-30 | X-ray | S7J, PO4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 21.61% | 27.40% | 9.60% | 2-(trifluoromethyl)pyrimidine-5-carboxamide; PHOSPHATE ION | 2-(trifluoromethyl)pyrimidine-5-carboxamide; PHOSPHATE ION | XDS | 97.10% | 6.6 | Diamond (I04-1) | REFMAC | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 6.0 | - | No | - | 17.2 | 25.7 | 31.2 | - | ||||||
| 5rm4 | 2.96 Å | 2020-09-30 | X-ray | PO4, PK4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 16.79% | 25.33% | 21.20% | PHOSPHATE ION; 2-fluoro-N,3-dimethylbenzene-1-sulfonamide | PHOSPHATE ION; 2-fluoro-N,3-dimethylbenzene-1-sulfonamide | XDS | 94.00% | 6.8 | Diamond (I04-1) | REFMAC | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 22.8 | - | No | - | 32.2 | 60.5 | 28.0 | - | ||||||
| 5rm5 | 2.06 Å | 2020-09-30 | X-ray | PO4, NUA | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 20.59% | 26.21% | 9.10% | PHOSPHATE ION; N-(1-ethyl-1H-pyrazol-4-yl)cyclobutanecarboxamide | PHOSPHATE ION; N-(1-ethyl-1H-pyrazol-4-yl)cyclobutanecarboxamide | XDS | 97.10% | 7.5 | Diamond (I04-1) | REFMAC | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 12.7 | - | No | - | 25.0 | 27.5 | 44.0 | - | ||||||
| 5rm6 | 2.13 Å | 2020-09-30 | X-ray | PO4, HR5 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 16.22% | 25.11% | 9.10% | PHOSPHATE ION; ~{N}-(cyclobutylmethyl)-1,5-dimethyl-pyrazole-4-carboxamide | PHOSPHATE ION; ~{N}-(cyclobutylmethyl)-1,5-dimethyl-pyrazole-4-carboxamide | XDS | 97.10% | 7.1 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 14.8 | - | No | - | 34.1 | 24.1 | 44.0 | - | ||||||
| 5rm7 | 1.84 Å | 2020-09-30 | X-ray | PO4, N0E | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 22.16% | 26.86% | 8.10% | PHOSPHATE ION; ~{N}-(4-hydroxyphenyl)-3-phenyl-propanamide | PHOSPHATE ION; ~{N}-(4-hydroxyphenyl)-3-phenyl-propanamide | XDS | 96.20% | 6.8 | Diamond (I04-1) | REFMAC | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 10.8 | - | No | - | 20.5 | 22.7 | 47.9 | - | ||||||
| 5rm8 | 2.14 Å | 2020-09-30 | X-ray | GQJ, PO4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 16.69% | 24.83% | 11.40% | methyl (2~{S},4~{R})-1-(furan-2-ylcarbonyl)-4-oxidanyl-pyrrolidine-2-carboxylate; PHOSPHATE ION | methyl (2~{S},4~{R})-1-(furan-2-ylcarbonyl)-4-oxidanyl-pyrrolidine-2-carboxylate; PHOSPHATE ION | XDS | 97.10% | 6.3 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 16.3 | - | No | - | 36.8 | 26.4 | 42.8 | - | ||||||
| 5rm9 | 2.08 Å | 2020-09-30 | X-ray | PO4, EJQ | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 16.71% | 25.43% | 7.60% | PHOSPHATE ION; ~{N}-(4-fluorophenyl)-2-pyrrolidin-1-yl-ethanamide | PHOSPHATE ION; ~{N}-(4-fluorophenyl)-2-pyrrolidin-1-yl-ethanamide | XDS | 96.60% | 8.0 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 12.1 | - | No | - | 31.4 | 19.4 | 44.1 | - | ||||||
| 5rma | 1.89 Å | 2020-09-30 | X-ray | PO4, JHJ | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 17.60% | 23.56% | 7.00% | PHOSPHATE ION; N-(4-methoxyphenyl)-N'-pyridin-4-ylurea | PHOSPHATE ION; N-(4-methoxyphenyl)-N'-pyridin-4-ylurea | XDS | 96.00% | 7.4 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 21.2 | - | No | - | 49.4 | 18.2 | 49.6 | - | ||||||
| 5rmb | 2.21 Å | 2020-09-30 | X-ray | VWV, PO4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 16.48% | 24.90% | 10.10% | ethyl (1,1-dioxo-1lambda~6~,4-thiazinan-4-yl)acetate; PHOSPHATE ION | ethyl (1,1-dioxo-1lambda~6~,4-thiazinan-4-yl)acetate; PHOSPHATE ION | XDS | 97.00% | 6.8 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 18.5 | - | No | - | 36.0 | 31.9 | 43.1 | - | ||||||
| 5rmc | 2.15 Å | 2020-09-30 | X-ray | PO4, 6SU | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 20.89% | 25.75% | 10.90% | PHOSPHATE ION; methyl 3-(methylsulfonylamino)benzoate | PHOSPHATE ION; methyl 3-(methylsulfonylamino)benzoate | XDS | 97.20% | 7.5 | Diamond (I04-1) | REFMAC | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 12.4 | - | No | - | 28.7 | 26.0 | 40.9 | - | ||||||
| 5rmd | 1.92 Å | 2020-09-30 | X-ray | VWY, PO4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 21.74% | 26.05% | 7.50% | N-ethyl-4-[(methylsulfonyl)amino]benzamide; PHOSPHATE ION | N-ethyl-4-[(methylsulfonyl)amino]benzamide; PHOSPHATE ION | XDS | 95.80% | 7.4 | Diamond (I04-1) | REFMAC | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 20.1 | - | No | - | 26.2 | 34.4 | 54.2 | - | ||||||
| 5rme | 2.23 Å | 2020-09-30 | X-ray | RYM, PO4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 21.18% | 28.44% | 12.40% | 4-(benzimidazol-1-ylmethyl)benzenecarbonitrile; PHOSPHATE ION | 4-(benzimidazol-1-ylmethyl)benzenecarbonitrile; PHOSPHATE ION | XDS | 96.20% | 6.3 | Diamond (I04-1) | REFMAC | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 8.3 | - | No | - | 12.1 | 45.1 | 25.5 | - | ||||||
| 5rmf | 2.23 Å | 2020-09-30 | X-ray | NX7, PO4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 20.75% | 27.04% | 12.10% | (2,6-difluorophenyl)(pyrrolidin-1-yl)methanone; PHOSPHATE ION | (2,6-difluorophenyl)(pyrrolidin-1-yl)methanone; PHOSPHATE ION | XDS | 97.20% | 6.2 | Diamond (I04-1) | REFMAC | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 20.4 | - | No | - | 19.3 | 57.2 | 38.8 | - | ||||||
| 5rmg | 2.12 Å | 2020-09-30 | X-ray | PO4, MUK | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 17.01% | 25.59% | 9.90% | PHOSPHATE ION; 4,6-dimethyl-~{N}-phenyl-pyrimidin-2-amine | PHOSPHATE ION; 4,6-dimethyl-~{N}-phenyl-pyrimidin-2-amine | XDS | 96.80% | 6.3 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 11.6 | - | No | - | 30.2 | 23.5 | 39.7 | - | ||||||
| 5rmh | 2.02 Å | 2020-09-30 | X-ray | VX4, PO4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 16.33% | 24.26% | 9.80% | [(4S)-4-methylazepan-1-yl](1,3-thiazol-4-yl)methanone; PHOSPHATE ION | [(4S)-4-methylazepan-1-yl](1,3-thiazol-4-yl)methanone; PHOSPHATE ION | XDS | 96.60% | 6.3 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 30.3 | - | No | - | 42.5 | 52.2 | 42.0 | - | ||||||
| 5rmi | 2.12 Å | 2020-09-30 | X-ray | STV, PO4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 16.43% | 25.13% | 11.50% | ~{N}-(1,3-benzodioxol-5-ylmethyl)ethanesulfonamide; PHOSPHATE ION | ~{N}-(1,3-benzodioxol-5-ylmethyl)ethanesulfonamide; PHOSPHATE ION | XDS | 97.10% | 6.1 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 17.9 | - | No | - | 34.0 | 27.1 | 48.6 | - | ||||||
| 5rmj | 2.10 Å | 2020-09-30 | X-ray | JOV, PO4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 23.30% | 29.65% | 9.40% | 3-chloro-N-(1-hydroxy-2-methylpropan-2-yl)benzamide; PHOSPHATE ION | 3-chloro-N-(1-hydroxy-2-methylpropan-2-yl)benzamide; PHOSPHATE ION | XDS | 96.70% | 6.8 | Diamond (I04-1) | REFMAC | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 2.2 | - | No | - | 8.1 | 16.2 | 30.9 | - | ||||||
| 5rmk | 2.08 Å | 2020-09-30 | X-ray | PO4, O2A | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 22.16% | 27.37% | 11.10% | PHOSPHATE ION; N-methyl-1H-indole-7-carboxamide | PHOSPHATE ION; N-methyl-1H-indole-7-carboxamide | XDS | 97.00% | 6.0 | Diamond (I04-1) | REFMAC | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 7.5 | - | No | - | 17.5 | 18.5 | 44.0 | - | ||||||
| 5rml | 2.43 Å | 2020-09-30 | X-ray | PO4, VXD | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 20.70% | 28.85% | 12.10% | PHOSPHATE ION; N-(3-chloro-2-methylphenyl)glycinamide | PHOSPHATE ION; N-(3-chloro-2-methylphenyl)glycinamide | XDS | 97.20% | 6.3 | Diamond (I04-1) | REFMAC | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 4.3 | - | No | - | 10.5 | 31.3 | 24.7 | - | ||||||
| 5rmm | 2.20 Å | 2020-09-30 | X-ray | PO4, VXG | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 21.23% | 28.05% | 12.10% | PHOSPHATE ION; (3S,4S)-1-acetyl-4-phenylpyrrolidine-3-carboxylic acid | PHOSPHATE ION; (3S,4S)-1-acetyl-4-phenylpyrrolidine-3-carboxylic acid | XDS | 97.30% | 6.7 | Diamond (I04-1) | BUSTER | 2020-09-16 | 34381037 | ZN | - | - | - | N/A | - | - | Helicase | 3.9 | - | No | - | 13.8 | 18.5 | 32.4 | - | ||||||
| 6zb4 | 3.03 Å | 2020-09-30 | Cryo-EM | NAG, EIC | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | LINOLEIC ACID; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose; LINOLEIC ACID | - | - | () | PHENIX | 2020-09-16 | 32958580 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 6zb5 | 2.85 Å | 2020-09-30 | Cryo-EM | NAG, EIC | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | LINOLEIC ACID; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose; LINOLEIC ACID | - | - | () | PHENIX | 2020-06-06 | 32958580 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7a92 | 4.20 Å | 2020-09-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-06-07 | 32942285 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7cwb | 1.90 Å | 2020-09-30 | XFEL | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 21.81% | 25.69% | N/A | - | CrystFEL | 99.91% | 10.4 | FREE ELECTRON LASER () | PHENIX | 2020-09-01 | 34403647 | - | - | http://covid19.bioreproducibility.org/static/data/7cwb/7cwb_refined.pdb | http://covid19.bioreproducibility.org/static/data/7cwb/7cwb_refined.mtz | N/A | - | According to the PDB validation report, more than half of the residues are RSRZ outliers. However, the map looks quite OK, and a quick run of Molprobity shows that the structure is in the top 96%. Interestingly, the title of the deposit talks about changes in the active site, but we were unable to find any. HOH 410 cannot be a water but a split of glu290 (too close) and Val 104 collides with HOH (bad rotamer), but those are minor abandonments. | 3C-like proteinase | N/A | minimal | Yes | - | N/A | N/A | N/A | - | |||||||
| 7cwc | 2.10 Å | 2020-09-30 | XFEL | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 23.07% | 25.93% | N/A | - | CrystFEL | 100.00% | 11.2 | FREE ELECTRON LASER () | PHENIX | 2020-08-27 | 34403647 | - | - | http://covid19.bioreproducibility.org/static/data/7cwc/7cwc_refined.pdb | http://covid19.bioreproducibility.org/static/data/7cwc/7cwc_refined.mtz | 25.50% | Placed the coordinates in a standardized location in the unit cell using ACHESYM. Added TLS. | Coordinates not in standardized location. | 3C-like proteinase | N/A | minimal | Yes | Yes | - | N/A | N/A | N/A | - | ||||||
| 7k3g | - | 2020-09-30 | NMR | - | SARS-CoV-2 | Envelope | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2020-08-27 | 33177698 | - | - | - | - | N/A | - | - | Envelope small membrane protein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7k3n | 1.65 Å | 2020-09-30 | X-ray | - | SARS-CoV-2 | NSP1 | No functional ligands | 20.30% | 24.82% | N/A | - | HKL-3000 | 99.80% | 35.6 | APS (19-ID) | PHENIX | 2020-09-11 | 33319167 | - | - | - | - | N/A | - | - | Host translation inhibitor nsp1 | 24.5 | - | No | - | 36.9 | 38.8 | 48.6 | - | |||||||
| 7k6d | 1.48 Å | 2020-09-30 | X-ray | SV6 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.47% | 21.53% | 13.50% | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide | XDS; Aimless; AutoProcess | 100.00% | 7.7 | NSLS-II (17-ID-1) | PHENIX | 2020-09-11 | 35842458 | - | - | http://covid19.bioreproducibility.org/static/data/7k6d/7k6d_refined.pdb | http://covid19.bioreproducibility.org/static/data/7k6d/7k6d_refined.mtz | 22.60% | Placed the coordinates in a standardized location in the unit cell using ACHESYM. | The coordinates are NOT in a standardized location in the unit cell. | 3C-like proteinase | 69.9 | minimal | Yes | Yes | - | 69.5 | 64.3 | 81.8 | - | |||||
| 7k6e | 1.63 Å | 2020-09-30 | X-ray | SV6 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 21.41% | 24.53% | 13.60% | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide | XDS; AutoProcess | 100.00% | 9.2 | NSLS-II (17-ID-1) | PHENIX | 2020-09-19 | 35842458 | - | - | http://covid19.bioreproducibility.org/static/data/7k6e/7k6e_refined.pdb | http://covid19.bioreproducibility.org/static/data/7k6e/7k6e_refined.mtz | 25.50% | Placed the coordinates in a standardized location in the unit cell using ACHESYM. | The coordinates are NOT in a standardized location in the unit cell. | 3C-like proteinase | 49.6 | minimal | Yes | Yes | - | 39.8 | 56.0 | 78.5 | - | |||||
| 7k7p | 1.77 Å | 2020-09-30 | X-ray | - | SARS-CoV-2 | NSP1 | No functional ligands | 18.84% | 21.65% | 7.10% | - | HKL-2000 | 99.81% | 40.38 | ROTATING ANODE () | PHENIX | 2020-09-19 | 33234675 | - | - | - | - | N/A | - | - | Non-structural protein 1 | 42.5 | - | No | - | 68.4 | 35.9 | 56.4 | - | |||||||
| 7d3i | 2.00 Å | 2020-09-30 | X-ray | GQU, TRS | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.59% | 20.89% | 6.30% | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL; (3~{S},3~{a}~{S},6~{a}~{R})-2-[3-[3,5-bis(fluoranyl)phenyl]propanoyl]-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide | (3~{S},3~{a}~{S},6~{a}~{R})-2-[3-[3,5-bis(fluoranyl)phenyl]propanoyl]-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide; 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL | XDS | 99.30% | 16.6 | PETRA III, DESY (P11) | BUSTER | 2020-09-23 | 33602867 | - | - | http://covid19.bioreproducibility.org/static/data/7d3i/7d3i_refined.pdb | http://covid19.bioreproducibility.org/static/data/7d3i/7d3i_refined.mtz | 20.70% | Achesymed, reoriented Glu288, added 6 waters. | The coordinates are NOT in a standardized location in the unit cell. | 3C-like proteinase | 83.1 | minimal | Yes | Yes | - | 75.1 | 93.3 | 86.7 | - | |||||
| 7d47 | 1.97 Å | 2020-10-07 | X-ray | CA | SARS-CoV-2 | NSP3: PLpro | No functional ligands | 18.75% | 19.73% | N/A | CALCIUM ION | CALCIUM ION | 98.81% | 19.55 | NSRRC (BL13B1) | PHENIX | 2020-09-19 | - | CA; ZN | - | http://covid19.bioreproducibility.org/static/data/7d47/7d47_rerefined.pdb | http://covid19.bioreproducibility.org/static/data/7d47/7d47_rerefined.mtz | 20.52% | Added 9 residues in chain B. The electron density in residues 188-195 and 227-230 is not very strong, but it was possible to trace the amino acids. The crystal is twinned. | The coordinates are NOT in a standardized location in the unit cell. Twinned crystal. Mising residues. | Non-structural protein 3 | 59.0 | significant | Yes | Yes | - | - | 83.5 | 69.3 | 39.8 | - | |||||
| 7joy | 2.00 Å | 2020-10-07 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 21.70% | 25.20% | 13.80% | - | xia2 | 96.30% | 3.9 | ROTATING ANODE () | BUSTER | 2020-09-22 | 33208735 | - | - | http://covid19.bioreproducibility.org/static/data/7joy/7joy_refined.pdb | http://covid19.bioreproducibility.org/static/data/7joy/7joy_refined.mtz | 28.50% | Reindexed -h, -k, -l to make it isomorphous with 7jox (replaced by 7khp). Placed the coordinates in a standardized location in the unit cell using ACHESYM. | The coordinates are NOT in a standardized location in the unit cell. | 3C-like proteinase | 21.5 | minimal | Yes | Yes | - | 33.3 | 29.5 | 55.1 | - | ||||||
| 7jp1 | 1.80 Å | 2020-10-07 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 18.90% | 23.25% | 6.40% | - | xia2 | 96.90% | 9.8 | APS (23-ID-B) | REFMAC | 2020-08-07 | 33208735 | - | - | http://covid19.bioreproducibility.org/static/data/7jp1/7jp1_refined.pdb | http://covid19.bioreproducibility.org/static/data/7jp1/7jp1_refined.mtz | 24.90% | Placed the coordinates in a standardized location in the unit cell using ACHESYM. | The coordinates are NOT in a standardized location in the unit cell. | 3C-like proteinase | 57.6 | minimal | Yes | Yes | - | 52.6 | 73.8 | 63.3 | - | ||||||
| 7jwb | 6.00 Å | 2020-10-07 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2020-08-07 | 33082574 | - | - | - | - | N/A | - | - | autonomous human heavy chain variable domain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7k43 | 2.60 Å | 2020-10-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-08-25 | 32972994 | - | - | - | - | N/A | - | - | SARS-CoV-2 spike glycoprotein; S2M11 Fab fragment (heavy chain); S2M11 Fab fragment (light chain) | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7k45 | 3.70 Å | 2020-10-07 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | 2020-09-14 | 32972994 | - | - | - | - | N/A | - | - | S2E12 neutralizing antibody Fab fragment (heavy chain); S2E12 neutralizing antibody Fab fragment (light chain); Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7k4n | 3.30 Å | 2020-10-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-09-14 | 32972994 | - | - | - | - | N/A | - | - | Spike glycoprotein; S2E12 neutralizing antibody Fab fragment (heavy chain); S2E12 neutralizing antibody Fab fragment (light chain) | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 6xkp | 2.72 Å | 2020-10-07 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 21.99% | 26.94% | 19.90% | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-3000 | 90.80% | 3.7 | SSRL (BL12-1) | REFMAC | 2020-09-15 | 33058755 | - | - | - | - | N/A | - | - | Spike protein S1; CV07-270 Heavy Chain; CV07-270 Light Chain | 13.7 | - | No | - | 19.9 | 38.9 | 40.6 | - | |||||||
| 6xkq | 2.55 Å | 2020-10-14 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 20.40% | 25.46% | 11.90% | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-3000 | 99.60% | 6.2 | SSRL (BL12-1) | PHENIX | 2020-06-26 | 33058755 | - | - | - | - | N/A | - | - | Spike protein S1; CV07-250 Heavy Chain; CV07-250 Light Chain | 22.2 | - | No | - | 31.2 | 31.6 | 56.6 | - | |||||||
| 6zpe | 1.58 Å | 2020-10-14 | X-ray | - | SARS-CoV-2 | NSP10 | No functional ligands | 15.15% | 15.98% | 4.40% | - | XDS | 99.90% | 24.1 | MAX IV (BioMAX) | PHENIX | 2020-06-26 | 33036230 | ZN | - | - | - | N/A | - | - | Replicase polyprotein 1ab | 64.5 | - | No | - | 97.1 | 25.0 | 81.7 | - | |||||||
| 7jmw | 2.89 Å | 2020-10-14 | X-ray | - | SARS-CoV-2 | Spike | Protein-protein complex | 23.79% | 29.08% | N/A | - | HKL-2000 | 97.90% | 7.4 | SSRL (BL12-1) | PHENIX; REFMAC | 2020-07-08 | 33242394 | - | - | - | - | N/A | - | - | Spike protein S1; COVA1-16 heavy chain; COVA1-16 light chain | 24.0 | - | No | - | 9.7 | 70.1 | 43.5 | - | |||||||
| 7jv2 | 3.50 Å | 2020-10-14 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2020-08-03 | 32991844 | - | - | - | - | N/A | - | - | S2H13 Fab heavy chain; S2H13 Fab light chain; SARS-CoV-2 spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7jv4 | 3.40 Å | 2020-10-14 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-08-20 | 32991844 | - | - | - | - | N/A | - | - | SARS-CoV-2 spike glycoprotein; S2H13 Fab heavy chain; S2H13 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7jv6 | 3.00 Å | 2020-10-14 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-08-20 | 32991844 | - | - | - | - | N/A | - | - | SARS-CoV-2 spike glycoprotein; S2H13 Fab heavy chain; S2H13 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7jva | 3.60 Å | 2020-10-14 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2020-08-20 | 32991844 | - | - | - | - | N/A | - | - | S2A4 Fab heavy chain; S2A4 Fab light chain; SARS-CoV-2 spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7jvc | 3.30 Å | 2020-10-14 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-08-20 | 32991844 | - | - | - | - | N/A | - | - | SARS-CoV-2 spike glycoprotein; S2A4 Fab heavy chain; S2A4 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7jw0 | 4.30 Å | 2020-10-14 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-08-20 | 32991844 | - | - | - | - | N/A | - | - | SARS-CoV-2 spike glycoprotein; S304 Fab light chain; S304 Fab heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7jx3 | 2.65 Å | 2020-10-14 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 25.78% | 28.81% | 19.90% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.60% | 8.7 | ALS (4.2.2) | REFMAC | 2020-08-24 | 32991844 | - | - | - | - | N/A | - | - | Light chain of Fab domain of monoclonal antibody S309; Heavy chain of Fab domain of monoclonal antibody S309; Light chain of Fab domain of monoclonal antibody S2H14; Heavy chain of Fab domain of monoclonal antibody S2H14; Light chain of Fab domain of monoclonal antibody S304; Heavy chain of Fab domain of monoclonal antibody S304; Spike glycoprotein - Receptor binding domain (UNP residues 328-531) | 54.3 | - | No | - | 10.6 | 78.5 | 94.3 | - | ||||||
| 7k5i | 2.90 Å | 2020-10-14 | Cryo-EM | - | SARS-CoV-2 | NSP1 | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2020-08-26 | 32995777 | MG; ZN | - | - | - | N/A | - | - | 40S ribosomal protein S17; 40S ribosomal protein S18; 40S ribosomal protein S19; 40S ribosomal protein S20; 40S ribosomal protein S25; 40S ribosomal protein S28; 40S ribosomal protein S27a; Receptor of activated protein C kinase 1; 40S ribosomal protein SA; 40S ribosomal protein S3a; 40S ribosomal protein S2; 40S ribosomal rRNA18S; 40S ribosomal protein S4, X isoform; 40S ribosomal protein S6; 40S ribosomal protein S7; 40S ribosomal protein S8; 40S ribosomal protein S9; 40S ribosomal protein S11; 40S ribosomal protein S13; 40S ribosomal protein S14; 40S ribosomal protein S21; 40S ribosomal protein S15a; 40S ribosomal protein S29; 40S ribosomal protein S23; 40S ribosomal protein S24; 40S ribosomal protein S26; 40S ribosomal protein S27; 40S ribosomal protein S30; 60S ribosomal protein L41; SARS-COV-2 Nsp1; 40S ribosomal protein S3; 40S ribosomal protein S5; 40S ribosomal protein S10; 40S ribosomal protein S12; 40S ribosomal protein S15; 40S ribosomal protein S16 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7kag | 3.21 Å | 2020-10-14 | X-ray | - | SARS-CoV-2 | NSP3: Ubl1 | No functional ligands | 21.43% | 24.78% | 5.30% | - | HKL-3000 | 96.00% | 17.53 | APS (19-ID) | PHENIX | 2020-09-16 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 - ubiquitin-like domain | 54.3 | - | No | - | - | 37.5 | 95.7 | 50.1 | - | ||||||
| 7a29 | 2.94 Å | 2020-10-14 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2020-09-30 | 33149112 | - | - | - | - | N/A | - | - | Spike glycoprotein; Neutralising sybody (Sb23) | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7k8m | 3.20 Å | 2020-10-21 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 17.53% | 23.42% | 16.00% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.70% | 6.0 | SSRL (BL12-1) | PHENIX | 2020-08-16 | 33045718 | - | - | - | - | N/A | - | - | C102 Fab Heavy Chain; C102 Fab Light Chain; Spike glycoprotein - receptor binding domain (UNP residues 331-517) | 50.3 | - | No | - | 50.8 | 93.3 | 31.7 | - | ||||||
| 7k8s | 3.40 Å | 2020-10-21 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-09-27 | 33045718 | - | - | - | - | N/A | - | - | Spike glycoprotein; C002 Fab Heavy Chain; C002 Fab Light Chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7k8t | 3.40 Å | 2020-10-21 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-09-27 | 33045718 | - | - | - | - | N/A | - | - | Spike glycoprotein; C002 Fab Heavy Chain; C002 Fab Light Chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7k8u | 3.80 Å | 2020-10-21 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-09-27 | 33045718 | - | - | - | - | N/A | - | - | Spike glycoprotein; C104 Fab Heavy Chain; C104 Fab Light Chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7k8v | 3.80 Å | 2020-10-21 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-09-27 | 33045718 | - | - | - | - | N/A | - | - | Spike glycoprotein; C110 Fab Heavy Chain; C110 Fab Light Chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7k8w | 3.60 Å | 2020-10-21 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-09-27 | 33045718 | - | - | - | - | N/A | - | - | Spike glycoprotein; C119 Fab Heavy Chain; C119 Fab Light Chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7k8x | 3.90 Å | 2020-10-21 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-09-27 | 33045718 | - | - | - | - | N/A | - | - | Spike glycoprotein; C121 Fab Heavy chain; C121 Fab Light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7k8y | 4.40 Å | 2020-10-21 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-09-27 | 33045718 | - | - | - | - | N/A | - | - | Spike glycoprotein; C121 Fab Heavy chain; C121 Fab Light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7k8z | 3.50 Å | 2020-10-21 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-09-27 | 33045718 | - | - | - | - | N/A | - | - | Spike glycoprotein; C135 Fab Heavy Chain; C135 Fab Light Chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7k90 | 3.24 Å | 2020-10-21 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-09-27 | 33045718 | - | - | - | - | N/A | - | - | Spike glycoprotein; C144 Fab Heavy Chain; C144 Fab Light Chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7k9p | 2.60 Å | 2020-10-21 | X-ray | - | SARS-CoV-2 | NSP15 | No functional ligands | 19.14% | 20.88% | N/A | - | CrystFEL | 100.00% | 3.1 | FREE ELECTRON LASER () | REFMAC | 2020-09-27 | 36630960 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 79.8 | - | No | - | 75.2 | 95.3 | 71.8 | - | |||||||
| 7kdt | 3.05 Å | 2020-10-21 | Cryo-EM | - | SARS-CoV-2 | ORF9B | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2020-09-29 | 33060197 | - | - | - | - | N/A | - | - | Mitochondrial import receptor subunit TOM70; ORF9b protein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7act | - | 2020-10-21 | NMR | - | SARS-CoV-2 | Nucleocapsid/RNA | Protein-protein complex | N/A | N/A | N/A | ssRNA | - | - | - | () | 2020-10-09 | 33264373 | - | - | - | - | N/A | - | A hybrid of experimental (protein) and modeling (nucleic acid) components | Nucleoprotein; ssRNA | N/A | hybrid | No | - | N/A | N/A | N/A | ssRNA | - | |||||||
| 7aku | 2.50 Å | 2020-10-28 | X-ray | RN2 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.23% | 23.53% | 19.00% | Calpeptin | Calpeptin | autoPROC | 86.23% | 3.712 | PETRA III, DESY (P11) | PHENIX | 2020-09-11 | 33811162 | - | - | - | - | N/A | - | - | Replicase polyprotein 1ab | 44.6 | - | No | - | 49.6 | 88.6 | 26.6 | - | ||||||
| 7d1m | 1.35 Å | 2020-10-28 | X-ray | K36 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 11.67% | 15.70% | 4.70% | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | HKL-2000 | 98.70% | 11.4 | SSRF (BL17U1) | REFMAC | 2020-10-02 | 32887884 | - | - | - | - | N/A | - | - | 3C-like proteinase | 66.9 | - | No | - | 97.5 | 39.8 | 71.6 | - | ||||||
| 7jjc | 2.36 Å | 2020-10-28 | X-ray | - | SARS-CoV-2 | Spike | Protein-protein complex | 20.32% | 24.99% | 10.00% | Spike glycoprotein | - | XDS | 99.60% | 11.4 | Australian Synchrotron (MX1) | PHENIX | 2020-09-14 | 33082294 | - | - | - | - | N/A | - | - | Neuropilin-1; Spike glycoprotein | 49.3 | - | No | - | 35.4 | 80.4 | 58.0 | - | ||||||
| 7jn5 | 2.71 Å | 2020-10-28 | X-ray | PO4, NAG | SARS-CoV | Spike | Protein-protein complex | 21.11% | 27.21% | 9.20% | PHOSPHATE ION; 2-acetamido-2-deoxy-beta-D-glucopyranose | PHOSPHATE ION; 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 97.50% | 6.7 | SSRL (BL12-2) | PHENIX | 2020-07-25 | 33275640 | - | - | - | - | N/A | - | - | CR3022 heavy chain; CR3022 light chain; Spike glycoprotein | 15.9 | - | No | - | 18.3 | 56.6 | 30.0 | - | ||||||
| 7k9z | 2.95 Å | 2020-10-28 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 26.12% | 28.71% | 16.80% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.90% | 12.3 | APS (23-ID-D) | PHENIX | 2020-08-03 | 34135340 | - | - | - | - | N/A | - | - | 52 Fab Heavy Chain; 52 Fab Light Chain; 298 Fab Light Chain; 298 Fab Heavy Chain; Spike glycoprotein | 14.7 | - | No | - | 11.0 | 49.9 | 40.9 | - | ||||||
| 7kfi | 1.60 Å | 2020-10-28 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 20.20% | 21.80% | 15.90% | - | XDS | 99.90% | 8.5 | LNLS SIRUS (MANACA) | BUSTER | 2020-09-29 | 34174328 | - | - | - | - | N/A | - | - | Main protease - UNP residues 3264-3569 | 47.4 | - | No | - | 66.9 | 15.6 | 87.5 | - | |||||||
| 7kg3 | 1.45 Å | 2020-10-28 | X-ray | MLI, MES | SARS-CoV-2 | NSP3: Macro | No functional ligands | 15.76% | 17.74% | 9.79% | MALONATE ION; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | MALONATE ION; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | XDS | 94.14% | 11.53 | () | PHENIX | 2020-10-14 | 33636189 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 85.2 | - | No | - | 93.1 | 89.6 | 84.1 | - | ||||||
| 7khp | 1.95 Å | 2020-10-28 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Protein-protein complex | 20.36% | 24.79% | 4.60% | - | xia2 | 99.60% | 10.8 | APS (23-ID-B) | REFMAC | 2020-10-15 | 33208735 | - | - | - | - | N/A | - | - | 3C-like proteinase | 12.5 | - | No | - | 37.4 | 12.6 | 45.8 | - | |||||||
| 7a4n | 2.75 Å | 2020-10-28 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-10-21 | 33431842 | - | - | - | - | N/A | - | - | Spike glycoprotein,Fibritin | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7acs | - | 2020-11-04 | NMR | - | SARS-CoV-2 | Nucleocapsid/RNA | Protein-protein complex | N/A | N/A | N/A | RNA (5'-R(P*GP*UP*CP*AP*GP*UP*G)-3'); RNA (5'-R(P*CP*AP*CP*UP*GP*AP*C)-3') | - | - | - | () | 2020-08-20 | 33264373 | - | - | - | - | N/A | - | A hybrid of experimental (protein) and modeling (nucleic acid) components | Nucleoprotein; RNA (5'-R(P*CP*AP*CP*UP*GP*AP*C)-3'); RNA (5'-R(P*GP*UP*CP*AP*GP*UP*G)-3') | N/A | hybrid | No | - | N/A | N/A | N/A | RNA (5'-R(P*CP*AP*CP*UP*GP*AP*C)-3'); RNA (5'-R(P*GP*UP*CP*AP*GP*UP*G)-3') | - | |||||||
| 7ad1 | 2.92 Å | 2020-11-04 | Cryo-EM | NAG | HIV-1/SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-09-11 | 33431842 | - | - | - | - | N/A | - | - | Spike glycoprotein,Envelope glycoprotein,Spike glycoprotein,Envelope glycoprotein,SARS-CoV-2 S protein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7cxm | 2.90 Å | 2020-11-04 | Cryo-EM | - | SARS-CoV-2 | NSP7/NSP8/NSP12/NSP13/RNA | Protein-protein complex | N/A | N/A | N/A | RNA (25-MER); RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3'); RNA (26-MER) | - | - | - | () | PHENIX | 2020-09-14 | 33208736 | ZN | - | - | - | N/A | - | - | nsp12 - UNP residues 4393-5324; nsp8 - UNP residues 3943-4140; nsp7 - UNP residues 3860-3942; RNA (25-MER); RNA (26-MER); RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3'); nsp13 - UNP residues 5325-5925 | N/A | - | No | - | N/A | N/A | N/A | RNA (25-MER); RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3'); RNA (26-MER) | - | ||||||
| 7cxn | 3.84 Å | 2020-11-04 | Cryo-EM | - | SARS-CoV-2 | NSP7/NSP8/NSP12/NSP13/RNA | Protein-protein complex | N/A | N/A | N/A | RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3'); Template RNA; Primer RNA | - | - | - | () | PHENIX | 2020-09-02 | 33208736 | ZN | - | - | - | N/A | - | - | nsp12 - UNP residues 4393-5324; nsp8 - UNP residues 3943-4140; nsp7 - UNP residues 3860-3942; Primer RNA; Template RNA; RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3'); nsp13 - UNP residues 5325-5925 | N/A | - | No | - | N/A | N/A | N/A | RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3'); Primer RNA; Template RNA | - | ||||||
| 7d6h | 1.60 Å | 2020-11-04 | X-ray | PO4 | SARS-CoV-2 | NSP3: PLpro | No functional ligands | 16.18% | 17.34% | 7.70% | PHOSPHATE ION | PHOSPHATE ION | XDS | 99.99% | 20.6 | SSRF (BL17U1) | PHENIX | 2020-09-02 | 33979649 | ZN | - | - | - | N/A | - | - | Papain-like protease | 79.6 | - | No | - | 94.3 | 50.1 | 97.1 | - | ||||||
| 7kdg | 3.01 Å | 2020-11-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-10-05 | 33417835 | - | - | - | - | N/A | - | - | Spike glycoprotein - ectodomain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7kdh | 3.33 Å | 2020-11-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-10-08 | 33417835 | - | - | - | - | N/A | - | - | Spike glycoprotein - ectodomain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7kdi | 3.26 Å | 2020-11-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-10-08 | 33417835 | - | - | - | - | N/A | - | - | Spike glycoprotein - ectodomain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7kdj | 3.49 Å | 2020-11-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-10-08 | 33417835 | - | - | - | - | N/A | - | - | Spike glycoprotein - ectodomain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7kdk | 2.80 Å | 2020-11-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-10-08 | 33417835 | - | - | - | - | N/A | - | - | Spike glycoprotein - ectodomain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7kdl | 2.96 Å | 2020-11-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-10-08 | 33417835 | - | - | - | - | N/A | - | - | Spike glycoprotein - ectodomain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ke4 | 3.21 Å | 2020-11-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-10-08 | 33417835 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ke6 | 3.10 Å | 2020-11-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-10-10 | 33417835 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ke7 | 3.32 Å | 2020-11-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-10-10 | 33417835 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ke8 | 3.26 Å | 2020-11-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-10-10 | 33417835 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ke9 | 3.08 Å | 2020-11-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-10-10 | 33417835 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7kea | 3.33 Å | 2020-11-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-10-10 | 33417835 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7keb | 3.48 Å | 2020-11-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-10-10 | 33417835 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7kec | 3.84 Å | 2020-11-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-10-10 | 33417835 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7c33 | 3.83 Å | 2020-11-04 | X-ray | APR | SARS-CoV-2 | NSP3: Macro | Functional ligand | 23.49% | 26.82% | 16.30% | ADENOSINE-5-DIPHOSPHORIBOSE | ADENOSINE-5-DIPHOSPHORIBOSE | HKL-2000 | 97.70% | 7.68 | NSRRC (TPS 05A) | PHENIX | 2020-10-10 | 32946224 | - | - | - | - | N/A | - | - | Macro domain from non-structural protein 3 | 40.7 | - | No | - | 20.7 | 89.6 | 46.9 | - | ||||||
| 7cjf | 2.11 Å | 2020-11-11 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 18.46% | 22.85% | 9.10% | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 92.70% | 7.9 | SSRF (BL17U) | PHENIX | 2020-05-11 | 33976198 | - | - | - | - | N/A | - | - | antibody heavy chain; antibody light chain; Spike glycoprotein - receptor binding domain, RBD | 47.3 | - | No | - | 56.6 | 56.9 | 56.3 | - | |||||||
| 7cz4 | 2.64 Å | 2020-11-11 | X-ray | APR | SARS-CoV-2 | NSP3: Macro | Functional ligand | 17.10% | 21.50% | 10.70% | ADENOSINE-5-DIPHOSPHORIBOSE | ADENOSINE-5-DIPHOSPHORIBOSE | HKL-2000 | 100.00% | 37.91 | NSRRC (TPS 05A) | PHENIX | 2020-07-10 | 32946224 | - | - | - | - | N/A | - | - | Macro domain from non-structural protein 3 | 71.6 | - | No | - | 69.7 | 93.6 | 56.0 | - | ||||||
| 7d4f | 2.57 Å | 2020-11-11 | Cryo-EM | H3U | SARS-CoV-2 | NSP7/NSP8/NSP12 | Functional ligand | N/A | N/A | N/A | 8-(3-(3-aminobenzamido)-4-methylbenzamido)naphthalene-1,3,5-trisulfonic acid | 8-(3-(3-aminobenzamido)-4-methylbenzamido)naphthalene-1,3,5-trisulfonic acid | - | - | () | PHENIX | 2020-09-07 | 33674802 | ZN | - | - | - | N/A | - | - | Non-structural protein 8; Non-structural protein 7; Non-structural protein 12 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7kip | 3.39 Å | 2020-11-11 | Cryo-EM | NAG | H-CoV-NL63 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-09-23 | 32817943 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7kj2 | 3.60 Å | 2020-11-11 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-10-24 | 33432247 | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7kj3 | 3.70 Å | 2020-11-11 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-10-25 | 33432247 | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7kj4 | 3.40 Å | 2020-11-11 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-10-25 | 33432247 | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7kj5 | 3.60 Å | 2020-11-11 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-10-25 | 33432247 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7kkk | 3.03 Å | 2020-11-11 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-10-25 | 33154106 | - | - | - | - | N/A | - | - | SARS-CoV-2 spike glycoprotein; Synthetic nanobody Nb6 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7kkl | 2.85 Å | 2020-11-11 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-10-27 | 33154106 | - | - | - | - | N/A | - | - | SARS-CoV-2 spike glycoprotein; Synthetic nanobody mNb6 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7kl9 | 4.10 Å | 2020-11-11 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-10-27 | 33154107 | - | - | - | - | N/A | - | - | Spike glycoprotein; CTC-445.2 inhibitor | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7cdi | 2.96 Å | 2020-11-11 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 20.20% | 25.38% | 15.50% | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.68% | 10.1 | SSRF (BL17U) | PHENIX | 2020-10-29 | 33431856 | - | - | - | - | N/A | - | - | Spike protein S1 - receptor binding domain; antibody P2C-1F11 heavy chain; antibody P2C-1F11 light chain | 30.3 | - | No | - | 31.9 | 69.1 | 35.5 | - | |||||||
| 7cdj | 3.40 Å | 2020-11-18 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 23.14% | 27.09% | 11.80% | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.74% | 8.8 | SSRF (BL17U) | PHENIX | 2020-06-19 | 33431856 | - | - | - | - | N/A | - | - | Spike protein S1 - receptor binding domain; antibody P2C-1A3 heavy chain; antibody P2C-1A3 light chain | 1.4 | - | No | - | 19.1 | 12.3 | 17.7 | - | |||||||
| 7ct5 | 4.00 Å | 2020-11-18 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-06-19 | 33177651 | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7kjr | 2.08 Å | 2020-11-18 | Cryo-EM | PEE | SARS-CoV-2 | ORF3a | Protein-protein complex | N/A | N/A | N/A | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine | - | - | () | PHENIX; phenix.real_space_refine | 2020-08-18 | 34158638 | - | - | - | - | N/A | - | - | Protein 3a; Apolipoprotein A-I - UNP residues 79-267 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7koa | 2.40 Å | 2020-11-18 | X-ray | GTA, SAM | SARS-CoV-2 | NSP10/NSP16 | Protein-protein complex | 23.39% | 27.66% | 12.90% | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE; S-ADENOSYLMETHIONINE | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE; S-ADENOSYLMETHIONINE | Precognition | 65.85% | 16.5 | APS (14-ID-B) | REFMAC | 2020-10-26 | - | ZN | - | http://covid19.bioreproducibility.org/static/data/7koa/7koa_refined.pdb | http://covid19.bioreproducibility.org/static/data/7koa/7koa_refined.mtz | 0.28% | Placed the coordinates in a standardized location in the unit cell using ACHESYM. | Two nice ligands and one with weak density (A303), but I would not remodel. Both Zn ions look very good. Weak density for B131-139, but nothing to fix. | 2'-O-methyltransferase; Non-structural protein 10 | 49.8 | minimal | Yes | Yes | - | - | 15.9 | 73.2 | 85.6 | - | ||||
| 7koj | 2.02 Å | 2020-11-18 | X-ray | Y94, MES, UNX | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 18.32% | 20.27% | 13.10% | 2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]-5-{[(prop-2-en-1-yl)carbamoyl]amino}benzamide; UNKNOWN ATOM OR ION; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | 2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]-5-{[(prop-2-en-1-yl)carbamoyl]amino}benzamide; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID; UNKNOWN ATOM OR ION | HKL-3000 | 99.90% | 6.1 | APS (19-ID) | REFMAC | 2020-11-07 | - | ZN | - | - | - | N/A | - | - | Papain-like protease | 41.3 | - | No | - | 79.9 | 17.6 | 61.0 | - | ||||||
| 7kok | 2.00 Å | 2020-11-18 | X-ray | Y96, MES, UNX | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 18.40% | 21.06% | 13.80% | 5-[(E)-(hydroxyimino)methyl]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID; UNKNOWN ATOM OR ION | 5-[(E)-(hydroxyimino)methyl]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID; UNKNOWN ATOM OR ION | HKL-3000 | 99.90% | 6.3 | APS (19-ID) | REFMAC | 2020-11-09 | - | ZN | - | - | - | N/A | - | - | Papain-like protease | 49.3 | - | No | - | 73.6 | 24.7 | 75.4 | - | ||||||
| 7kol | 2.58 Å | 2020-11-18 | X-ray | Y96, MES | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 18.75% | 20.61% | 9.80% | 5-[(E)-(hydroxyimino)methyl]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | 5-[(E)-(hydroxyimino)methyl]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | HKL-3000 | 100.00% | 6.8 | APS (19-ID) | REFMAC | 2020-11-09 | - | ZN | - | - | - | N/A | - | - | Papain-Like Protease | 51.3 | - | No | - | 77.2 | 29.8 | 70.5 | - | ||||||
| 7kph | 1.46 Å | 2020-11-18 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 16.70% | 18.09% | 4.90% | - | XDS | 94.90% | 12.8 | LNLS SIRUS (MANACA) | PHENIX | 2020-11-09 | 34174328 | - | - | - | - | N/A | - | - | 3C-like proteinase | 79.0 | - | No | - | 91.8 | 78.7 | 68.8 | - | |||||||
| 5s6x | 2.32 Å | 2020-11-18 | X-ray | WUG | SARS-CoV-2 | NSP15 | PanDDA fragment screening | 18.37% | 22.18% | 29.10% | 1-(2,4-dimethyl-1H-imidazol-5-yl)methanamine | 1-(2,4-dimethyl-1H-imidazol-5-yl)methanamine | XDS | 100.00% | 6.6 | Diamond (I04-1) | BUSTER | 2020-11-11 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 74.4 | - | No | - | 63.4 | 87.7 | 75.2 | - | ||||||
| 5s6y | 2.32 Å | 2020-11-25 | X-ray | WUJ | SARS-CoV-2 | NSP15 | PanDDA fragment screening | 20.93% | 25.37% | 31.00% | N-[(furan-2-yl)methyl]urea | N-[(furan-2-yl)methyl]urea | XDS | 100.00% | 5.9 | Diamond (I04-1) | BUSTER | 2020-11-13 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 58.3 | - | No | - | 32.0 | 87.7 | 71.4 | - | ||||||
| 5s6z | 2.28 Å | 2020-11-25 | X-ray | WUM | SARS-CoV-2 | NSP15 | PanDDA fragment screening | 19.09% | 22.23% | 45.10% | 4-[(dimethylamino)methyl]-1,3-thiazol-2-amine | 4-[(dimethylamino)methyl]-1,3-thiazol-2-amine | XDS | 100.00% | 5.0 | Diamond (I04-1) | BUSTER | 2020-11-13 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 71.6 | - | No | - | 62.8 | 79.0 | 77.4 | - | ||||||
| 5s70 | 2.33 Å | 2020-11-25 | X-ray | WUS | SARS-CoV-2 | NSP15 | PanDDA fragment screening | 18.20% | 22.76% | 29.20% | (5R)-2-methyl-4,5,6,7-tetrahydro-1H-benzimidazol-5-amine | (5R)-2-methyl-4,5,6,7-tetrahydro-1H-benzimidazol-5-amine | XDS | 99.90% | 6.6 | Diamond (I04-1) | BUSTER | 2020-11-13 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 70.3 | - | No | - | 57.5 | 83.2 | 75.6 | - | ||||||
| 5s71 | 1.94 Å | 2020-11-25 | X-ray | WUV | SARS-CoV-2 | NSP15 | PanDDA fragment screening | 18.40% | 21.47% | 21.40% | 5'-thiothymidine | 5'-thiothymidine | XDS | 100.00% | 6.4 | Diamond (I04-1) | BUSTER | 2020-11-13 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 73.6 | - | No | - | 70.1 | 79.0 | 75.1 | - | ||||||
| 5s72 | 2.51 Å | 2020-11-25 | X-ray | WUY | SARS-CoV-2 | NSP15 | PanDDA fragment screening | 21.10% | 27.67% | 38.70% | N-(2-aminoethyl)-N'-phenylurea | N-(2-aminoethyl)-N'-phenylurea | XDS | 99.60% | 4.5 | Diamond (I04-1) | BUSTER | 2020-11-13 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 38.3 | - | No | - | 15.8 | 90.0 | 46.9 | - | ||||||
| 7a25 | 3.06 Å | 2020-11-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-11-13 | 33149112 | - | - | - | - | N/A | - | - | Spike glycoprotein; Sybody 23 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7ddd | 3.00 Å | 2020-11-25 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | PHENIX | 2020-08-16 | 33431876 | - | - | - | - | N/A | - | - | Spike glycoprotein - UNP residues 1-1208 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ddn | 6.30 Å | 2020-11-25 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | PHENIX | 2020-10-28 | 33431876 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7jwy | 2.50 Å | 2020-11-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-10-29 | 33271067 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 5rl0 | 1.69 Å | 2020-11-25 | X-ray | VEG | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.74% | 21.40% | 9.70% | ethyl N-[(2R)-2-[(4-tert-butylphenyl)(propanoyl)amino]-2-(pyridin-3-yl)acetyl]-beta-alaninate | ethyl N-[(2R)-2-[(4-tert-butylphenyl)(propanoyl)amino]-2-(pyridin-3-yl)acetyl]-beta-alaninate | XDS | 99.40% | 7.4 | Diamond (I04-1) | BUSTER | 2020-08-26 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 68.0 | - | No | - | - | 70.6 | 55.9 | 84.8 | - | |||||
| 5rl1 | 1.65 Å | 2020-12-02 | X-ray | VEJ | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.19% | 21.03% | 11.70% | N-(4-tert-butylphenyl)-N-[(1R)-2-[(3-methoxypropyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | N-(4-tert-butylphenyl)-N-[(1R)-2-[(3-methoxypropyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | XDS | 98.80% | 6.1 | Diamond (I04-1) | BUSTER | 2020-08-05 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 70.6 | - | No | - | - | 73.8 | 73.6 | 69.6 | - | |||||
| 5rl2 | 1.48 Å | 2020-12-02 | X-ray | VEM | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.81% | 19.99% | 7.30% | N-(4-tert-butylphenyl)-N-[(1R)-2-[(2-methoxyethyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | N-(4-tert-butylphenyl)-N-[(1R)-2-[(2-methoxyethyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | XDS | 95.60% | 7.9 | Diamond (I04-1) | BUSTER | 2020-08-05 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 64.8 | - | No | - | - | 81.8 | 48.6 | 73.9 | - | |||||
| 5rl3 | 1.51 Å | 2020-12-02 | X-ray | VEP | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.71% | 20.49% | 7.50% | N-(4-tert-butylphenyl)-N-[(1R)-2-[(oxan-4-yl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | N-(4-tert-butylphenyl)-N-[(1R)-2-[(oxan-4-yl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | XDS | 99.00% | 7.4 | Diamond (I04-1) | BUSTER | 2020-08-05 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 66.0 | - | No | - | - | 78.3 | 48.6 | 80.0 | - | |||||
| 5rl4 | 1.53 Å | 2020-12-02 | X-ray | VEV | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.95% | 20.52% | 8.40% | N-(4-tert-butylphenyl)-N-[(1R)-2-(methylamino)-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | N-(4-tert-butylphenyl)-N-[(1R)-2-(methylamino)-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | XDS | 98.70% | 7.1 | Diamond (I04-1) | BUSTER | 2020-08-05 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 62.9 | - | No | - | - | 77.9 | 48.6 | 73.9 | - | |||||
| 5rl5 | 1.58 Å | 2020-12-02 | X-ray | VEY | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.42% | 21.74% | 7.50% | N-(4-tert-butylphenyl)-N-[(1R)-2-(ethylamino)-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | N-(4-tert-butylphenyl)-N-[(1R)-2-(ethylamino)-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | XDS | 99.50% | 7.5 | Diamond (I04-1) | BUSTER | 2020-08-05 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 70.0 | - | No | - | - | 67.6 | 55.9 | 92.2 | - | |||||
| 6z5t | 1.57 Å | 2020-12-02 | X-ray | APR, NA | SARS-CoV-2 | NSP3: Macro | Functional ligand | 20.94% | 24.93% | N/A | SODIUM ION; ADENOSINE-5-DIPHOSPHORIBOSE | ADENOSINE-5-DIPHOSPHORIBOSE; SODIUM ION | xia2 | 100.00% | 4.1 | Diamond (I03) | PHENIX | 2020-08-05 | 33202171 | - | - | - | - | N/A | - | - | Replicase polyprotein 1ab | 53.5 | - | No | - | 35.8 | 81.2 | 64.8 | - | ||||||
| 6z6i | 2.00 Å | 2020-12-02 | X-ray | A1R, NA, MPO | SARS-CoV-2 | NSP3: Macro | Functional ligand | 22.89% | 27.86% | 29.90% | 3[N-MORPHOLINO]PROPANE SULFONIC ACID; SODIUM ION; 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE; SODIUM ION; 3[N-MORPHOLINO]PROPANE SULFONIC ACID | xia2 | 99.90% | 2.6 | Diamond (I03) | PHENIX | 2020-05-27 | 33202171 | - | - | - | - | N/A | - | - | Replicase polyprotein 1ab | 27.9 | - | No | - | 14.8 | 64.1 | 52.6 | - | ||||||
| 6z72 | 2.30 Å | 2020-12-02 | X-ray | A3R, NA, MLT | SARS-CoV-2 | NSP3: Macro | Functional ligand | 21.80% | 26.32% | 16.40% | D-MALATE; Adenosine Diphosphate (Hydroxymethyl)pyrrolidine monoalcohol | Adenosine Diphosphate (Hydroxymethyl)pyrrolidine monoalcohol; SODIUM ION; D-MALATE | xia2 | 99.90% | 3.2 | Diamond (I03) | PHENIX | 2020-05-28 | 33202171 | - | - | - | - | N/A | - | - | Replicase polyprotein 1ab | 50.2 | - | No | - | 24.2 | 81.2 | 70.2 | - | ||||||
| 7a1u | 1.67 Å | 2020-12-02 | X-ray | FUA, IMD | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.28% | 20.42% | 7.50% | FUSIDIC ACID; IMIDAZOLE | FUSIDIC ACID; IMIDAZOLE | DIALS | 99.87% | 8.5 | PETRA III, DESY (P11) | REFMAC; PHENIX | 2020-05-29 | 33811162 | - | - | - | - | N/A | - | - | Main Protease | 70.5 | - | No | - | 78.7 | 56.0 | 82.2 | - | ||||||
| 7abu | 1.60 Å | 2020-12-02 | X-ray | R6Q, IMD | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.66% | 21.51% | N/A | IMIDAZOLE; 1'-[2-[4-(trifluoromethyl)phenyl]ethyl]spiro[1~{H}-3,1-benzoxazine-4,4'-piperidine]-2-one | 1'-[2-[4-(trifluoromethyl)phenyl]ethyl]spiro[1~{H}-3,1-benzoxazine-4,4'-piperidine]-2-one; IMIDAZOLE | XDS; DIALS | 99.60% | 5.85 | PETRA III, DESY (P11) | PHENIX | 2020-08-14 | 33811162 | - | - | - | - | N/A | - | - | Main Protease | 76.7 | - | No | - | 69.6 | 83.7 | 79.2 | - | ||||||
| 7adw | 1.63 Å | 2020-12-02 | X-ray | R7Q, IMD | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.63% | 22.87% | 9.50% | 2-methyl-1-(4-methylphenyl)propan-1-one; IMIDAZOLE | 2-methyl-1-(4-methylphenyl)propan-1-one; IMIDAZOLE | XDS | 97.60% | 7.6 | PETRA III, DESY (P11) | PHENIX | 2020-09-08 | 33811162 | - | - | - | - | N/A | - | - | Main Protease | 63.2 | - | No | - | 56.4 | 73.6 | 71.2 | - | ||||||
| 7af0 | 1.70 Å | 2020-12-02 | X-ray | R9W | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.76% | 22.50% | N/A | 2,3,5,6,7,8-hexahydro-1~{H}-cyclopenta[b]quinolin-9-amine | 2,3,5,6,7,8-hexahydro-1~{H}-cyclopenta[b]quinolin-9-amine | XDS | 99.73% | 6.05 | PETRA III, DESY (P11) | REFMAC | 2020-09-16 | 33811162 | - | - | - | - | N/A | - | - | Main Protease | 63.7 | - | No | - | 60.1 | 60.2 | 81.8 | - | ||||||
| 7aga | 1.68 Å | 2020-12-02 | X-ray | LZE | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.58% | 22.25% | N/A | 4-{[(2,6-dichlorophenyl)carbonyl]amino}-N-piperidin-4-yl-1H-pyrazole-3-carboxamide | 4-{[(2,6-dichlorophenyl)carbonyl]amino}-N-piperidin-4-yl-1H-pyrazole-3-carboxamide | XDS | 97.80% | 9.01 | PETRA III, DESY (P11) | PHENIX; REFMAC | 2020-09-18 | 33811162 | - | - | - | - | N/A | - | - | Main Protease | 74.2 | - | No | - | 62.7 | 78.7 | 84.5 | - | ||||||
| 7aha | 1.68 Å | 2020-12-02 | X-ray | SIN | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.55% | 20.11% | 7.20% | SUCCINIC ACID | SUCCINIC ACID | DIALS | 96.70% | 9.5 | PETRA III, DESY (P11) | PHENIX; REFMAC | 2020-09-22 | 33811162 | - | - | - | - | N/A | - | - | Main Protease | 83.5 | - | No | - | 80.9 | 83.7 | 92.6 | - | ||||||
| 7ak4 | 1.63 Å | 2020-12-02 | X-ray | CB1 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.33% | 22.05% | N/A | 5-(AZIRIDIN-1-YL)-2,4-DINITROBENZAMIDE | 5-(AZIRIDIN-1-YL)-2,4-DINITROBENZAMIDE | XDS | 98.50% | 6.79 | PETRA III, DESY (P11) | MAIN | 2020-09-24 | 33811162 | - | - | - | - | N/A | - | - | Main Protease | 39.0 | - | No | - | 64.6 | 45.6 | 43.7 | - | ||||||
| 7alh | 1.65 Å | 2020-12-02 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 16.47% | 18.90% | 5.60% | - | XDS | 99.70% | 12.3 | ELETTRA (11.2C) | PHENIX; PHENIX | 2020-09-29 | 34769210 | - | - | - | - | N/A | - | - | Main Protease | 82.9 | - | No | - | 88.2 | 68.7 | 97.1 | - | |||||||
| 7ali | 1.65 Å | 2020-12-02 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 18.88% | 21.67% | 6.90% | - | XDS | 98.40% | 9.8 | ELETTRA (11.2C) | PHENIX; PHENIX | 2020-10-06 | 34769210 | - | - | - | - | N/A | - | - | Main Protease | 71.7 | - | No | - | 68.3 | 62.2 | 89.1 | - | |||||||
| 7amj | 1.59 Å | 2020-12-02 | X-ray | RMZ | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.30% | 21.04% | 9.20% | (3~{S})-3-[2-[4-(3,4-dimethylphenyl)piperazin-1-yl]ethyl]-2,3-dihydroisoindol-1-one | (3~{S})-3-[2-[4-(3,4-dimethylphenyl)piperazin-1-yl]ethyl]-2,3-dihydroisoindol-1-one | DIALS | 99.80% | 6.4 | PETRA III, DESY (P11) | PHENIX; REFMAC | 2020-10-06 | 33811162 | - | - | - | - | N/A | - | - | Main Protease | 78.9 | - | No | - | 73.7 | 73.6 | 91.9 | - | ||||||
| 7ans | 1.70 Å | 2020-12-02 | X-ray | RNW | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.35% | 21.14% | N/A | 2-[(diphenylmethyl)-oxidanyl-$l^{3}-sulfanyl]-~{N}-oxidanyl-ethanamide | 2-[(diphenylmethyl)-oxidanyl-$l^{3}-sulfanyl]-~{N}-oxidanyl-ethanamide | DIALS | 99.70% | 7.51 | PETRA III, DESY (P11) | PHENIX | 2020-10-09 | 33811162 | - | - | - | - | N/A | - | - | Main Protease | 67.8 | - | No | - | 72.9 | 56.1 | 81.9 | - | ||||||
| 7aol | 1.47 Å | 2020-12-02 | X-ray | IMD, RQH | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.54% | 19.24% | N/A | IMIDAZOLE; (1~{S})-1-(4-chloranylphenoxy)-1-imidazol-1-yl-3,3-dimethyl-butan-2-one | IMIDAZOLE; (1~{S})-1-(4-chloranylphenoxy)-1-imidazol-1-yl-3,3-dimethyl-butan-2-one | DIALS; XDS | 99.67% | 14.61 | PETRA III, DESY (P11) | PHENIX; REFMAC | 2020-10-12 | 33811162 | - | - | - | - | N/A | - | - | ORF1ab polyprotein | 67.7 | - | No | - | 86.5 | 68.9 | 55.3 | - | ||||||
| 7ap6 | 1.78 Å | 2020-12-02 | X-ray | RQN | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.78% | 23.66% | N/A | 4-(4-ethyl-5-fluoranyl-2-oxidanyl-phenoxy)-3-fluoranyl-benzamide | 4-(4-ethyl-5-fluoranyl-2-oxidanyl-phenoxy)-3-fluoranyl-benzamide | DIALS | 99.70% | 4.46 | PETRA III, DESY (P11) | PHENIX | 2020-10-14 | 33811162 | - | - | - | - | N/A | - | - | Main Protease | 52.2 | - | No | - | 48.3 | 64.4 | 66.6 | - | ||||||
| 7aph | 1.65 Å | 2020-12-02 | X-ray | RT2 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 23.45% | 26.63% | N/A | Tofogliflozin | Tofogliflozin | XDS | 96.00% | 5.36 | PETRA III, DESY (P11) | MAIN | 2020-10-16 | 33811162 | - | - | - | - | N/A | - | - | Main Protease | 11.1 | - | No | - | 22.0 | 29.4 | 40.3 | - | ||||||
| 7aqi | 1.70 Å | 2020-12-02 | X-ray | QEL, IMD | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 22.60% | 26.56% | N/A | IMIDAZOLE; 4-[(1R,2S)-2-(4-benzylpiperidin-1-yl)-1-hydroxypropyl]phenol | 4-[(1R,2S)-2-(4-benzylpiperidin-1-yl)-1-hydroxypropyl]phenol; IMIDAZOLE | XDS | 98.49% | 4.35 | PETRA III, DESY (P11) | REFMAC | 2020-10-16 | 33811162 | - | - | - | - | N/A | - | - | Main Protease | 40.7 | - | No | - | 22.6 | 83.7 | 51.0 | - | ||||||
| 7aqj | 2.59 Å | 2020-12-02 | X-ray | S7H, RV8 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 21.74% | 25.96% | N/A | Triglycidyl isocyanurate; 1-[(2~{R})-2-oxidanylpropyl]-3-[[(2~{R})-oxiran-2-yl]methyl]-5-[[(2~{S})-oxiran-2-yl]methyl]-1,3,5-triazinane-2,4,6-trione | 1-[(2~{R})-2-oxidanylpropyl]-3-[[(2~{R})-oxiran-2-yl]methyl]-5-[[(2~{S})-oxiran-2-yl]methyl]-1,3,5-triazinane-2,4,6-trione; Triglycidyl isocyanurate | DIALS | 99.40% | 4.95 | PETRA III, DESY (P11) | PHENIX | 2020-10-21 | 33811162 | - | - | - | - | N/A | - | - | Main Protease | 34.6 | - | No | - | 26.9 | 42.6 | 75.7 | - | ||||||
| 7ar5 | 1.40 Å | 2020-12-02 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 17.57% | 21.37% | N/A | - | XDS | 95.50% | 10.2 | PETRA III, DESY (P11) | PHENIX | 2020-10-22 | 33811162 | - | - | - | - | N/A | - | - | SARS coronavirus 2 main proteinase | 65.7 | - | No | - | 70.9 | 55.9 | 79.4 | - | |||||||
| 7ar6 | 1.40 Å | 2020-12-02 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 16.53% | 19.04% | N/A | - | XDS | 99.50% | 11.56 | PETRA III, DESY (P11) | PHENIX | 2020-10-23 | 33811162 | - | - | - | - | N/A | - | - | Main Protease | 80.9 | - | No | - | 87.5 | 73.6 | 84.9 | - | |||||||
| 7arf | 2.00 Å | 2020-12-02 | X-ray | RVW | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.50% | 25.27% | 5.90% | (2~{S},3~{R},4~{R},5~{S},6~{S})-2-(hydroxymethyl)-6-sulfanyl-oxane-3,4,5-triol | (2~{S},3~{R},4~{R},5~{S},6~{S})-2-(hydroxymethyl)-6-sulfanyl-oxane-3,4,5-triol | XDS | 99.50% | 12.0 | PETRA III, DESY (P11) | PHENIX | 2020-10-23 | 33811162 | - | - | - | - | N/A | - | - | Main Protease | 53.4 | - | No | - | 32.9 | 73.4 | 75.3 | - | ||||||
| 7avd | 1.80 Å | 2020-12-02 | X-ray | S1W | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.42% | 23.87% | 15.70% | 3-[[5-[3-(dimethylamino)phenoxy]pyrimidin-2-yl]amino]phenol | 3-[[5-[3-(dimethylamino)phenoxy]pyrimidin-2-yl]amino]phenol | XDS | 99.50% | 5.6 | PETRA III, DESY (P11) | PHENIX | 2020-10-24 | 33811162 | - | - | - | - | N/A | - | - | Main Protease | 48.5 | - | No | - | 46.2 | 60.2 | 65.8 | - | ||||||
| 7awr | 1.34 Å | 2020-12-02 | X-ray | S7W | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 15.19% | 19.21% | N/A | TEGAFUR | TEGAFUR | DIALS | 98.88% | 11.78 | PETRA III, DESY (P11) | PHENIX | 2020-11-05 | 33811162 | - | - | - | - | N/A | - | - | Main Protease | 55.4 | - | No | - | 86.6 | 21.1 | 77.7 | - | ||||||
| 7aws | 1.81 Å | 2020-12-02 | X-ray | S8E | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.13% | 23.85% | 12.00% | 5-[[(2-bromoethylamino)-(ethylamino)phosphoryl]oxymethyl]-1-methyl-~{N},~{N}-bis(oxidanyl)imidazol-2-amine | 5-[[(2-bromoethylamino)-(ethylamino)phosphoryl]oxymethyl]-1-methyl-~{N},~{N}-bis(oxidanyl)imidazol-2-amine | DIALS | 100.00% | 5.0 | PETRA III, DESY (P11) | PHENIX | 2020-11-09 | 33811162 | - | - | - | - | N/A | - | - | Main Protease | 67.3 | - | No | - | 46.4 | 73.6 | 89.7 | - | ||||||
| 7awu | 2.07 Å | 2020-12-02 | X-ray | S8B | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 21.46% | 25.84% | N/A | ~{N}-propan-2-yl-5-(2-pyridin-4-ylethynyl)pyridine-2-carboxamide | ~{N}-propan-2-yl-5-(2-pyridin-4-ylethynyl)pyridine-2-carboxamide | DIALS | 99.80% | 4.29 | PETRA III, DESY (P11) | PHENIX | 2020-11-09 | 33811162 | - | - | - | - | N/A | - | - | Replicase polyprotein 1ab | 37.5 | - | No | - | 27.9 | 56.0 | 67.2 | - | ||||||
| 7aww | 1.65 Å | 2020-12-02 | X-ray | CLU | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.36% | 21.98% | N/A | 2,6-DICHLORO-N-IMIDAZOLIDIN-2-YLIDENEANILINE | 2,6-DICHLORO-N-IMIDAZOLIDIN-2-YLIDENEANILINE | DIALS | 97.40% | 12.5 | PETRA III, DESY (P11) | REFMAC; PHENIX | 2020-11-09 | 33811162 | - | - | - | - | N/A | - | - | Main Protease | 63.8 | - | No | - | 65.4 | 73.8 | 63.3 | - | ||||||
| 7ax6 | 1.95 Å | 2020-12-02 | X-ray | S8H, IMD | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.73% | 24.85% | N/A | (2~{S})-2-azanyl-5-oxidanylidene-5-[[(2~{S})-1-oxidanylidene-1-[(2-oxidanylidene-2-propan-2-yloxy-ethyl)amino]-3-sulfanyl-propan-2-yl]amino]pentanoic acid; IMIDAZOLE | (2~{S})-2-azanyl-5-oxidanylidene-5-[[(2~{S})-1-oxidanylidene-1-[(2-oxidanylidene-2-propan-2-yloxy-ethyl)amino]-3-sulfanyl-propan-2-yl]amino]pentanoic acid; IMIDAZOLE | DIALS | 98.27% | 10.27 | PETRA III, DESY (P11) | PHENIX | 2020-11-09 | 33811162 | - | - | - | - | N/A | - | - | Main Protease | 50.2 | - | No | - | 36.7 | 78.7 | 60.0 | - | ||||||
| 7axm | 1.40 Å | 2020-12-02 | X-ray | 93J, IMD | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 14.28% | 20.95% | N/A | (2E)-N-{4-[(3-chloro-4-fluorophenyl)amino]-3-cyano-7-ethoxyquinolin-6-yl}-4-(dimethylamino)but-2-enamide | (2E)-N-{4-[(3-chloro-4-fluorophenyl)amino]-3-cyano-7-ethoxyquinolin-6-yl}-4-(dimethylamino)but-2-enamide; IMIDAZOLE | DIALS | 98.59% | 10.07 | PETRA III, DESY (P11) | PHENIX; PHENIX | 2020-11-09 | 33811162 | - | - | - | - | N/A | - | - | Main Protease | 58.3 | - | No | - | 74.5 | 37.5 | 79.1 | - | ||||||
| 7axo | 1.65 Å | 2020-12-02 | X-ray | QCP | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.75% | 20.05% | N/A | AR-42 | AR-42 | XDS | 87.82% | 11.82 | PETRA III, EMBL c/o DESY (P14 (MX2)) | PHENIX | 2020-11-09 | 33811162 | - | - | - | - | N/A | - | - | Main Protease | 74.7 | - | No | - | 81.4 | 52.3 | 93.5 | - | ||||||
| 7ay7 | 1.55 Å | 2020-12-02 | X-ray | S8T, IMD | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.62% | 19.50% | N/A | 9-fluoranyl-3-propan-2-yl-5,6-dihydrobenzo[b][1]benzothiepine; IMIDAZOLE | 9-fluoranyl-3-propan-2-yl-5,6-dihydrobenzo[b][1]benzothiepine; IMIDAZOLE | DIALS | 98.96% | 13.75 | PETRA III, DESY (P11) | REFMAC; PHENIX | 2020-11-10 | 33811162 | - | - | - | - | N/A | - | - | Main Protease | 78.5 | - | No | - | 84.9 | 83.7 | 69.2 | - | ||||||
| 7dcc | 4.30 Å | 2020-12-02 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | PHENIX | 2020-11-11 | 33431876 | - | - | - | - | N/A | - | - | The heavy chain of 3C1 fab; SARC-CoV2 S protein; The light chain of 3C1 fab | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7dcx | 5.90 Å | 2020-12-02 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2020-10-24 | 33431876 | - | - | - | - | N/A | - | - | The heavy chain of 3C1 fab that binds with the up RBD; SARS-Cov2 S protein; The light chain of 3C1 fab | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7dd2 | 5.60 Å | 2020-12-02 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | PHENIX | 2020-10-27 | 33431876 | - | - | - | - | N/A | - | - | SARS-Cov2-S protein; The heavy chain of 3C1 fab; The light chain of 3C1 fab | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7dd8 | 7.50 Å | 2020-12-02 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | PHENIX | 2020-10-27 | 33431876 | - | - | - | - | N/A | - | - | The heavy chain of 3C1 fab; The light chain of 3C1 chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7dk4 | 3.80 Å | 2020-12-02 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2020-10-28 | 33431876 | - | - | - | - | N/A | - | - | The heavy chain of 2H2 Fab; The light chain of 2H2 Fab; SARS-CoV-2 S protein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7dk5 | 13.50 Å | 2020-12-02 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2020-11-23 | 33431876 | - | - | - | - | N/A | - | - | SARS-CoV-2 S protein; The heavy chain of 2H2 Fab; The light chain of 2H2 Fab | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7dk6 | 4.30 Å | 2020-12-02 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2020-11-23 | 33431876 | - | - | - | - | N/A | - | - | SARS-CoV-2 S protein; The heavy chain of 2H2 Fab; The light chain of 2H2 Fab | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7dk7 | 9.70 Å | 2020-12-02 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2020-11-23 | 33431876 | - | - | - | - | N/A | - | - | The heavy chain fragment of 2H2 Fab; The light chain fragment of 2H2 Fab; SARS-CoV-2 S protein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7jqb | 2.70 Å | 2020-12-02 | Cryo-EM | - | SARS-CoV-2 | NSP1 | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | PHENIX | 2020-11-23 | 33188728 | - | - | - | - | N/A | - | - | rRNA; uS7; eS31; eS6; RACK1; eS7; eS8; uS4; S10_plectin domain-containing protein; eS12; uS19; eS25; Uncharacterized protein; eS17; uS13; Uncharacterized protein; uS10; 40S ribosomal protein S21; 40S ribosomal protein S24; Nsp1 - UNP residues 145-180; 40S ribosomal protein S4; uS17; 40S ribosomal protein SA; uS15; Uncharacterized protein; uS8; Uncharacterized protein; eS27; 40S ribosomal protein S26; eS1; uS5; eS28; Ribosomal protein S3; eS30 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7jqc | 3.30 Å | 2020-12-02 | Cryo-EM | - | SARS-CoV-2 | NSP1 | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | PHENIX | 2020-08-10 | 33188728 | - | - | - | - | N/A | - | - | rRNA; uS7; eS31; eS6; RACK1; eS7; eS8; uS4; S10_plectin domain-containing protein; eS12; uS19; eS25; Uncharacterized protein; eS17; uS13; Uncharacterized protein; uS10; 40S ribosomal protein S21; 40S ribosomal protein S24; Nsp1 - UNP residues 145-180; Cricket paralysis virus IRES RNA; 40S ribosomal protein S4; 40S ribosomal protein SA; uS17; uS15; Uncharacterized protein; uS8; Uncharacterized protein; eS27; 40S ribosomal protein S26; eS1; uS5; eS28; Ribosomal protein S3; eS30 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7jvb | 3.29 Å | 2020-12-02 | X-ray | CAC | SARS-CoV-2 | Spike | Functional ligand | 28.13% | 32.22% | 15.60% | CACODYLATE ION | CACODYLATE ION | HKL-2000 | 96.34% | 7.8 | APS (23-ID-B) | PHENIX | 2020-08-10 | 33154108 | - | - | - | - | N/A | - | - | Spike glycoprotein; Nanobody Nb20 | 11.1 | - | No | - | 4.8 | 54.7 | 32.2 | - | ||||||
| 7keg | 2.90 Å | 2020-12-02 | X-ray | PO4 | SARS-CoV-2 | NSP15 | Functional ligand | 17.50% | 21.80% | 57.20% | PHOSPHATE ION | PHOSPHATE ION | XDS | 99.10% | 5.0 | LNLS SIRUS (MANACA) | BUSTER | 2020-08-20 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 55.8 | - | No | - | 66.9 | 70.3 | 49.0 | - | ||||||
| 7keh | 2.59 Å | 2020-12-02 | X-ray | B3P | SARS-CoV-2 | NSP15 | Functional ligand | 18.70% | 22.00% | 103.10% | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL | XDS | 94.80% | 7.6 | MAX IV (BioMAX) | BUSTER | 2020-10-10 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 43.1 | - | No | - | 65.0 | 48.9 | 48.1 | - | ||||||
| 7kf4 | 2.61 Å | 2020-12-02 | X-ray | - | SARS-CoV-2 | NSP15 | No functional ligands | 21.90% | 24.60% | N/A | - | XDS | 99.73% | 8.24 | MAX IV (BioMAX) | BUSTER | 2020-10-10 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 51.7 | - | No | - | 39.0 | 81.2 | 58.2 | - | |||||||
| 7kfv | 2.10 Å | 2020-12-02 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 18.30% | 21.55% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 98.90% | 8.98 | APS (24-ID-E) | BUSTER | 2020-10-13 | 33200128 | - | - | - | - | N/A | - | - | Spike glycoprotein; Heavy chain of antibody C1A-B12 Fab; light chain of antibody C1A-B3 Fab | 64.0 | - | No | - | 69.4 | 51.8 | 81.6 | - | ||||||
| 7kfw | 2.79 Å | 2020-12-02 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 18.93% | 22.93% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 98.10% | 5.41 | APS (24-ID-E) | BUSTER | 2020-10-15 | 33200128 | - | - | - | - | N/A | - | - | Spike glycoprotein; heavy chain of antibody C1A-B3 Fab; light chain of antibody C1A-B3 Fab | 34.6 | - | No | - | 55.7 | 36.6 | 52.9 | - | ||||||
| 7kfx | 2.23 Å | 2020-12-02 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 19.09% | 22.61% | 13.60% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.10% | 6.86 | APS (24-ID-C) | BUSTER | 2020-10-15 | 33200128 | - | - | - | - | N/A | - | - | Spike glycoprotein; heavy chain of human antibody C1A-C2 Fab; light chain of human antibody C1A-C2 Fab | 56.1 | - | No | - | 59.0 | 50.2 | 77.6 | - | ||||||
| 7kfy | 2.16 Å | 2020-12-02 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 20.54% | 24.86% | 11.80% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 94.00% | 6.17 | APS (24-ID-C) | BUSTER | 2020-10-15 | 33200128 | - | - | - | - | N/A | - | - | Spike glycoprotein; heavy chain of human antibody C1A-F10 Fab; light chain of human antibody C1A-F10 Fab | 50.1 | - | No | - | 36.6 | 55.3 | 83.2 | - | ||||||
| 7krx | 2.72 Å | 2020-12-02 | X-ray | Y41, UNX | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 20.97% | 24.90% | 17.30% | UNKNOWN ATOM OR ION; 3-amino-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide | 3-amino-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide; UNKNOWN ATOM OR ION | HKL-3000 | 100.00% | 6.3 | APS (19-ID) | REFMAC | 2020-10-15 | - | ZN | - | - | - | N/A | - | - | Papain-like protease | 43.7 | - | No | - | - | 36.0 | 64.7 | 62.2 | - | |||||
| 7cyq | 2.83 Å | 2020-12-02 | Cryo-EM | GDP | SARS-CoV-2 | NSP7/NSP8/NSP9/NSP12/NSP13 | Functional ligand | N/A | N/A | N/A | GUANOSINE-5'-DIPHOSPHATE; Template; Primer | GUANOSINE-5'-DIPHOSPHATE | - | - | () | PHENIX | 2020-11-20 | 33232691 | ZN; MG | - | - | - | N/A | - | - | Replicase polyprotein 1ab - UNP residues 4393-5324; nsp8 - UNP residues 3943-4140; nsp7 - UNP residues 3860-3942; Primer; Template; Replicase polyprotein 1ab - UNP residues 5325-5925; Replicase polyprotein 1ab - UNP residues 4141-4253 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7k0r | 3.30 Å | 2020-12-09 | Cryo-EM | U5P, PO4 | SARS-CoV-2 | NSP15 | Functional ligand | N/A | N/A | N/A | PHOSPHATE ION; URIDINE-5'-MONOPHOSPHATE | URIDINE-5'-MONOPHOSPHATE; PHOSPHATE ION | - | - | () | phenix.real_space_refine; PHENIX | 2020-09-04 | 33504779 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7kmb | 3.39 Å | 2020-12-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-09-04 | 33271067 | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7kms | 3.64 Å | 2020-12-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-11-02 | 33271067 | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; SARS-CoV-2 spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7kmz | 3.62 Å | 2020-12-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-11-03 | 33271067 | - | - | - | - | N/A | - | - | SARS-CoV-2 spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7knb | 3.93 Å | 2020-12-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-11-03 | 33271067 | - | - | - | - | N/A | - | - | SARS-CoV-2 spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7kqo | 0.85 Å | 2020-12-09 | X-ray | - | SARS-CoV-2 | NSP3: Macro | No functional ligands | 12.03% | 13.84% | 4.30% | - | XDS | 99.80% | 13.58 | ALS (8.3.1) | PHENIX | 2020-11-04 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 80.1 | - | No | - | 99.2 | 67.4 | 76.7 | - | |||||||
| 7kqp | 0.88 Å | 2020-12-09 | X-ray | AR6 | SARS-CoV-2 | NSP3: Macro | Functional ligand | 10.76% | 12.33% | 5.70% | [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | XDS | 98.90% | 13.26 | NSLS-II (17-ID-2) | PHENIX | 2020-11-17 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 73.4 | - | No | - | 99.8 | 43.7 | 80.4 | - | ||||||
| 7kqw | 0.93 Å | 2020-12-09 | X-ray | - | SARS-CoV-2 | NSP3: Macro | No functional ligands | 12.76% | 14.70% | 3.50% | - | XDS | 91.90% | 8.44 | NSLS-II (17-ID-2) | PHENIX | 2020-11-17 | 33853786 | - | - | - | - | N/A | - | - | NSP3 - macrodomain (UNP residues 1024-1192) | 67.2 | - | No | - | 98.6 | 34.4 | 76.6 | - | |||||||
| 7kr0 | 0.77 Å | 2020-12-09 | X-ray | - | SARS-CoV-2 | NSP3: Macro | No functional ligands | 10.99% | 11.72% | 11.70% | - | XDS | 93.00% | 31.52 | ALS (8.3.1) | PHENIX | 2020-11-17 | 33853786 | - | - | http://covid19.bioreproducibility.org/static/data/7kr0/7kr0_refined.pdb | http://covid19.bioreproducibility.org/static/data/7kr0/7kr0_refined.mtz | 0.12% | Differences: - 2 GLU, 3 VAL first 2 visible residues are split at the main chain and go in 2 different ways - there is some evidence for this but this part is heavily disordered - 5 SER - they have 3 conformations - no see map for the third - 4 ASN to 5 PHE - main chain and side chains are split - no evidence - 12 LEU - 15 ASN - main chain and side chains are split - no evidence - 14 ASP - split - possible but map is weak - 20 ASN - side chain split very weak evidence - 25 GLU - side chain split some evidence - 28 LYS to 32 PRO - main chain split - without it is not possible to split 28 and 29 - 28 LYS - side chain split weak - 29 LYS - side chain split weak - 30 VAL - side chain split weak - 31 LYS - side chain split no evidence - 40 ASN - side chain split some evidence - 42 TYR - side chain split some evidence - with one and occ 1.0 Refmac already increased B-factors to 50 and there is positive density - 44 LYS - side chain split some evidence - 55 LYS - side chain split some evidence - 58 ASN - side chain split some evidence - 64 GLU - side chain split no evidence - 66 ASP - side chain split some evidence but 2 conformations are close and in coot refinement I can not separate them - 66 ASP and 67 ASP - main chain split - no evidence - 72 ASN - split - no evidence - 75 LEU - side chain split weak map - 76 LYS - side chain split in 3 parts, new model has 2 - some evidence - 77 VAL - split - no evidence - 75 LEU to 77 VAL - main chain split - no evidence - 82 VAL - side chain split no evidence - 83 LEU - side chain split no evidence - 86 HIS - side chain split some evidence - 88 LEU - side chain split some evidence - 86 HIS to 88 LEU - main chain split - no evidence maybe required for 88? - 100 VAL to 103 GLY - good evidence for main chain split - 103 to 111 - main chain split - no evidence - 108 GLU - side chain split no evidence - 119 HIS - side chain split no evidence - 126 LEU - main chain split (side chain split in both versions) - weak - 133 to 137 - good evidence for main chain split - except 133 GLY - 136 PRO - side chain split - requires main chain split - 141 ARG - side chain split - some evidence - 154 to 159 - main chain split - no evidence except for 156 that we also split - 156 PHE - split into 2 not 3 - 162 - 165 - main chain split - no evidence - 166 - 168 - good evidence for main chain split - 170 GLU - side chain split weak | alternative conformations with poor density fit | NSP3 - macrodomain (UNP residues 1024-1192) | 85.2 | minimal | Yes | Yes | - | 99.9 | 95.7 | 71.1 | - | ||||||
| 7kr1 | 1.55 Å | 2020-12-09 | X-ray | - | SARS-CoV-2 | NSP3: Macro | No functional ligands | 17.97% | 22.14% | 19.32% | - | DIALS | 99.99% | 9.07 | ALS (8.3.1) | PHENIX | 2020-11-18 | 33853786 | - | - | - | - | N/A | - | - | NSP3 - macrodomain (UNP residues 1024-1192) | 57.8 | - | No | - | 63.8 | 63.1 | 63.3 | - | |||||||
| 7kvl | 2.09 Å | 2020-12-09 | X-ray | X4P, SER | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.12% | 22.72% | 16.10% | 2-chloropyridine-4-carboxamide; SERINE | 2-chloropyridine-4-carboxamide; SERINE | XDS | 96.80% | 10.0 | LNLS SIRUS (MANACA) | PHENIX | 2020-11-20 | 34174328 | - | - | - | - | N/A | - | - | Main protease - UNP residues 3264-3569 | 46.2 | - | No | - | 57.9 | 53.6 | 56.2 | - | ||||||
| 7kvr | 2.12 Å | 2020-12-09 | X-ray | X4V | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.00% | 22.45% | 10.10% | N~4~,N~4~-dimethylpyridine-2,4-diamine | N~4~,N~4~-dimethylpyridine-2,4-diamine | XDS | 97.30% | 12.2 | LNLS SIRUS (MANACA) | PHENIX | 2020-11-28 | 34174328 | - | - | - | - | N/A | - | - | Main protease - UNP residues 3264-3569 | 32.7 | - | No | - | 60.7 | 40.7 | 40.0 | - | ||||||
| 5rs7 | 1.00 Å | 2020-12-09 | X-ray | W4Y | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 18.01% | 19.00% | 4.20% | 1-{2-[(propan-2-yl)oxy]ethyl}-2-sulfanylidene-1,2,3,5-tetrahydro-4H-pyrrolo[3,2-d]pyrimidin-4-one | 1-{2-[(propan-2-yl)oxy]ethyl}-2-sulfanylidene-1,2,3,5-tetrahydro-4H-pyrrolo[3,2-d]pyrimidin-4-one | XDS | 99.90% | 16.51 | ALS (8.3.1) | PHENIX | 2020-11-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 51.6 | - | No | - | 87.7 | 19.4 | 70.9 | - | ||||||
| 5rs8 | 1.01 Å | 2020-12-16 | X-ray | H35 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.35% | 17.88% | 7.30% | N-(FURAN-2-YLMETHYL)-7H-PURIN-6-AMINE | N-(FURAN-2-YLMETHYL)-7H-PURIN-6-AMINE | XDS | 99.40% | 8.75 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 74.1 | - | No | - | 92.6 | 49.1 | 83.8 | - | ||||||
| 5rs9 | 1.00 Å | 2020-12-16 | X-ray | W4V | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.39% | 17.84% | 3.40% | 6,7-dihydro-5H-cyclopenta[d][1,2,4]triazolo[1,5-a]pyrimidin-8-amine | 6,7-dihydro-5H-cyclopenta[d][1,2,4]triazolo[1,5-a]pyrimidin-8-amine | XDS | 99.20% | 20.26 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 70.9 | - | No | - | 92.7 | 38.5 | 86.5 | - | ||||||
| 5rsb | 1.00 Å | 2020-12-16 | X-ray | W4S | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.69% | 18.39% | 3.70% | 7-methyl-7H-pyrrolo[2,3-d]pyrimidin-4-amine | 7-methyl-7H-pyrrolo[2,3-d]pyrimidin-4-amine | XDS | 99.50% | 18.76 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 73.1 | - | No | - | 90.7 | 46.2 | 86.1 | - | ||||||
| 5rsc | 1.01 Å | 2020-12-16 | X-ray | W5S | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 18.53% | 20.12% | 4.40% | 7-[(furan-2-yl)methyl]-5,6-dimethyl-7H-pyrrolo[2,3-d]pyrimidin-4-amine | 7-[(furan-2-yl)methyl]-5,6-dimethyl-7H-pyrrolo[2,3-d]pyrimidin-4-amine | XDS | 99.70% | 15.68 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 58.7 | - | No | - | 80.9 | 31.0 | 80.1 | - | ||||||
| 5rsd | 1.00 Å | 2020-12-16 | X-ray | 1LQ | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.03% | 18.87% | 5.20% | quinazolin-4-amine | quinazolin-4-amine | XDS | 99.00% | 13.13 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 64.1 | - | No | - | 88.4 | 32.6 | 81.8 | - | ||||||
| 5rse | 1.00 Å | 2020-12-16 | X-ray | W5P | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 19.13% | 20.84% | 3.40% | 4-[(3R)-3-fluoropiperidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine | 4-[(3R)-3-fluoropiperidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine | XDS | 99.90% | 19.31 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 44.7 | - | No | - | 75.4 | 16.5 | 73.1 | - | ||||||
| 5rsf | 1.00 Å | 2020-12-16 | X-ray | W5M | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.89% | 17.35% | 3.90% | 9-methyl-9H-purine-2,6-diamine | 9-methyl-9H-purine-2,6-diamine | XDS | 99.30% | 17.95 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 72.6 | - | No | - | 94.2 | 41.0 | 86.5 | - | ||||||
| 5rsg | 1.00 Å | 2020-12-16 | X-ray | W5J | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.78% | 18.26% | 3.80% | N-methyl-N-7H-pyrrolo[2,3-d]pyrimidin-4-yl-beta-alanine | N-methyl-N-7H-pyrrolo[2,3-d]pyrimidin-4-yl-beta-alanine | XDS | 99.50% | 17.75 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 59.0 | - | No | - | 91.2 | 29.3 | 72.1 | - | ||||||
| 5rsh | 1.00 Å | 2020-12-16 | X-ray | W5G | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.48% | 17.91% | 3.80% | 4-(5-azaspiro[2.5]octan-5-yl)-7H-pyrrolo[2,3-d]pyrimidine | 4-(5-azaspiro[2.5]octan-5-yl)-7H-pyrrolo[2,3-d]pyrimidine | XDS | 99.30% | 18.72 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 66.7 | - | No | - | 92.4 | 31.0 | 85.0 | - | ||||||
| 5rsi | 1.01 Å | 2020-12-16 | X-ray | W5D | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 19.84% | 21.69% | 6.70% | 4-(1,4-oxazonan-4-yl)-7H-pyrrolo[2,3-d]pyrimidine | 4-(1,4-oxazonan-4-yl)-7H-pyrrolo[2,3-d]pyrimidine | XDS | 99.50% | 9.11 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 30.9 | - | No | - | 68.2 | 15.8 | 53.9 | - | ||||||
| 5rsj | 1.00 Å | 2020-12-16 | X-ray | W5A | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.99% | 19.28% | 7.90% | 3-[(2-methyl-1,3-thiazol-4-yl)methyl]-3H-purin-6-amine | 3-[(2-methyl-1,3-thiazol-4-yl)methyl]-3H-purin-6-amine | XDS | 99.20% | 8.0 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 77.5 | - | No | - | 86.3 | 59.3 | 89.2 | - | ||||||
| 5rsk | 1.00 Å | 2020-12-16 | X-ray | W57 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.36% | 16.40% | 3.90% | 3-[(3-methoxy-1,2-oxazol-5-yl)methyl]-3H-purin-6-amine | 3-[(3-methoxy-1,2-oxazol-5-yl)methyl]-3H-purin-6-amine | XDS | 99.90% | 19.07 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 73.9 | - | No | - | 96.4 | 41.0 | 87.6 | - | ||||||
| 5rsl | 1.00 Å | 2020-12-16 | X-ray | W51 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.14% | 18.89% | 4.90% | 6-[(1s,4s)-2-azabicyclo[2.2.2]octan-2-yl]-5-chloropyrimidin-4-amine | 6-[(1s,4s)-2-azabicyclo[2.2.2]octan-2-yl]-5-chloropyrimidin-4-amine | XDS | 99.50% | 14.16 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 52.7 | - | No | - | 88.4 | 21.5 | 70.4 | - | ||||||
| 5rsm | 1.02 Å | 2020-12-16 | X-ray | 4SO | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.97% | 18.47% | 8.50% | 4-sulfamoylbenzoic acid | 4-sulfamoylbenzoic acid | XDS | 99.80% | 9.08 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 68.2 | - | No | - | 90.3 | 31.0 | 90.5 | - | ||||||
| 5rsn | 1.00 Å | 2020-12-16 | X-ray | 51X | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.64% | 18.07% | 9.20% | (1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)acetic acid | (1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)acetic acid | XDS | 98.70% | 7.44 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 75.4 | - | No | - | 91.9 | 41.0 | 95.9 | - | ||||||
| 5rso | 1.03 Å | 2020-12-16 | X-ray | TYZ | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.96% | 18.66% | 6.30% | PARA ACETAMIDO BENZOIC ACID | PARA ACETAMIDO BENZOIC ACID | XDS | 99.90% | 9.94 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 76.5 | - | No | - | 89.5 | 52.3 | 90.2 | - | ||||||
| 5rsp | 1.02 Å | 2020-12-16 | X-ray | LSA | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.82% | 17.22% | 6.50% | 1,2-BENZISOTHIAZOL-3(2H)-ONE 1,1-DIOXIDE | 1,2-BENZISOTHIAZOL-3(2H)-ONE 1,1-DIOXIDE | XDS | 99.70% | 9.84 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 78.1 | - | No | - | 94.6 | 49.1 | 93.0 | - | ||||||
| 5rsq | 1.00 Å | 2020-12-16 | X-ray | MOK | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.67% | 16.99% | 5.10% | 5-methyl-3-phenyl-1,2-oxazole-4-carboxylic acid | 5-methyl-3-phenyl-1,2-oxazole-4-carboxylic acid | XDS | 98.40% | 14.24 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 73.0 | - | No | - | 95.2 | 38.5 | 89.2 | - | ||||||
| 5rsr | 1.00 Å | 2020-12-16 | X-ray | XIY | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.25% | 19.13% | 6.90% | 2-HYDROXYMETHYL-BENZOIMIDAZOLE | 2-HYDROXYMETHYL-BENZOIMIDAZOLE | XDS | 97.10% | 8.82 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 76.2 | - | No | - | 87.1 | 52.3 | 91.8 | - | ||||||
| 5rss | 1.00 Å | 2020-12-16 | X-ray | NC3 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.30% | 16.66% | 7.40% | N-[(CYCLOHEXYLAMINO)CARBONYL]GLYCINE | N-[(CYCLOHEXYLAMINO)CARBONYL]GLYCINE | XDS | 96.50% | 9.15 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 82.3 | - | No | - | 95.9 | 67.2 | 88.4 | - | ||||||
| 5rst | 1.00 Å | 2020-12-16 | X-ray | 5HN | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.35% | 16.94% | 5.90% | 5-hydroxypyridine-3-carboxylic acid | 5-hydroxypyridine-3-carboxylic acid | XDS | 99.30% | 12.64 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 75.4 | - | No | - | 95.3 | 49.1 | 84.5 | - | ||||||
| 5rsu | 1.00 Å | 2020-12-16 | X-ray | OHB | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.97% | 16.33% | 7.40% | salicylamide | salicylamide | XDS | 98.60% | 9.83 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 78.1 | - | No | - | 96.5 | 55.7 | 84.6 | - | ||||||
| 5rsv | 1.03 Å | 2020-12-16 | X-ray | 4MB | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.26% | 17.95% | 5.40% | 4-[(METHYLSULFONYL)AMINO]BENZOIC ACID | 4-[(METHYLSULFONYL)AMINO]BENZOIC ACID | XDS | 99.90% | 12.33 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 64.1 | - | No | - | 92.3 | 31.0 | 79.7 | - | ||||||
| 5rsw | 1.00 Å | 2020-12-16 | X-ray | 6FZ | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.24% | 17.74% | 5.50% | 2,3-dihydro-1H-indene-2-carboxylic acid | 2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.10% | 12.24 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 74.7 | - | No | - | 93.1 | 43.4 | 90.5 | - | ||||||
| 5rsx | 1.00 Å | 2020-12-16 | X-ray | YTX | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.06% | 17.60% | 5.30% | 2-(3-methoxy-4-oxidanyl-phenyl)ethanoic acid | 2-(3-methoxy-4-oxidanyl-phenyl)ethanoic acid | XDS | 97.10% | 13.55 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 71.3 | - | No | - | 93.5 | 32.6 | 92.6 | - | ||||||
| 5rsy | 1.04 Å | 2020-12-16 | X-ray | 3R6 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.21% | 19.15% | 9.30% | 2-hydroxy-5-(methylsulfanyl)benzoic acid | 2-hydroxy-5-(methylsulfanyl)benzoic acid | XDS | 99.20% | 6.44 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 80.2 | - | No | - | 87.0 | 67.2 | 89.2 | - | ||||||
| 5rsz | 1.02 Å | 2020-12-16 | X-ray | ZZA | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.55% | 18.17% | 5.50% | 1-PHENYL-1H-PYRAZOLE-4-CARBOXYLIC ACID | 1-PHENYL-1H-PYRAZOLE-4-CARBOXYLIC ACID | XDS | 98.90% | 12.52 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 81.8 | - | No | - | 91.5 | 67.2 | 90.9 | - | ||||||
| 5rt0 | 1.00 Å | 2020-12-16 | X-ray | 4BL | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.38% | 15.67% | 5.70% | 6-methyl-1H-indole-2-carboxylic acid | 6-methyl-1H-indole-2-carboxylic acid | XDS | 96.40% | 13.99 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 80.9 | - | No | - | 97.6 | 59.3 | 89.2 | - | ||||||
| 5rt1 | 1.00 Å | 2020-12-16 | X-ray | 3A9 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.91% | 16.27% | 4.60% | 2,3-dihydro-1-benzofuran-5-carboxylic acid | 2,3-dihydro-1-benzofuran-5-carboxylic acid | XDS | 97.20% | 14.89 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 79.8 | - | No | - | 96.6 | 59.3 | 86.1 | - | ||||||
| 5rt2 | 1.00 Å | 2020-12-16 | X-ray | 5OF | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.73% | 17.44% | 4.90% | 2-(4-oxidanylidene-3~{H}-phthalazin-1-yl)ethanoic acid | 2-(4-oxidanylidene-3~{H}-phthalazin-1-yl)ethanoic acid | XDS | 97.10% | 13.9 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 74.4 | - | No | - | 94.0 | 46.2 | 86.1 | - | ||||||
| 5rt3 | 1.05 Å | 2020-12-16 | X-ray | 2FX | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 19.55% | 21.86% | 9.10% | 1-benzothiophen-2-ylacetic acid | 1-benzothiophen-2-ylacetic acid | XDS | 99.80% | 6.59 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 63.7 | - | No | - | 66.5 | 52.3 | 83.4 | - | ||||||
| 5rt4 | 1.02 Å | 2020-12-16 | X-ray | 4BX | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.00% | 17.52% | 9.30% | 3-(1H-benzimidazol-2-yl)propanoic acid | 3-(1H-benzimidazol-2-yl)propanoic acid | XDS | 97.90% | 8.58 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 66.8 | - | No | - | 93.7 | 24.4 | 90.5 | - | ||||||
| 5rt5 | 1.00 Å | 2020-12-16 | X-ray | 07L | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.34% | 17.15% | 7.00% | 7-hydroxy-2H-chromen-2-one | 7-hydroxy-2H-chromen-2-one | XDS | 96.80% | 10.01 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 76.4 | - | No | - | 94.7 | 55.7 | 81.3 | - | ||||||
| 5rt6 | 1.00 Å | 2020-12-16 | X-ray | 05R | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.72% | 18.17% | 5.30% | 2-(3,4-dichlorophenyl)ethanoic acid | 2-(3,4-dichlorophenyl)ethanoic acid | XDS | 97.60% | 14.58 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 67.8 | - | No | - | 91.5 | 36.4 | 83.0 | - | ||||||
| 5rt7 | 1.00 Å | 2020-12-16 | X-ray | GVH | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.89% | 17.35% | 10.00% | 1H-PYRROLO[2,3-B]PYRIDINE | 1H-PYRROLO[2,3-B]PYRIDINE | XDS | 97.10% | 6.61 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 79.7 | - | No | - | 94.2 | 55.7 | 91.8 | - | ||||||
| 5rt8 | 1.00 Å | 2020-12-16 | X-ray | HLR | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.75% | 15.99% | 6.20% | 1,2-benzoxazol-3-amine | 1,2-benzoxazol-3-amine | XDS | 97.50% | 12.35 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 79.9 | - | No | - | 97.1 | 52.3 | 93.0 | - | ||||||
| 5rt9 | 1.01 Å | 2020-12-16 | X-ray | 54G | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.05% | 18.52% | 5.50% | 2-hydroxy-5-methylbenzoic acid | 2-hydroxy-5-methylbenzoic acid | XDS | 98.40% | 11.84 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 76.9 | - | No | - | 90.1 | 52.3 | 90.5 | - | ||||||
| 5rta | 1.00 Å | 2020-12-16 | X-ray | Q6T | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.07% | 16.28% | 6.20% | 1,3-benzodioxole-4-carboxylic acid | 1,3-benzodioxole-4-carboxylic acid | XDS | 98.10% | 11.72 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 81.4 | - | No | - | 96.6 | 59.3 | 91.8 | - | ||||||
| 5rtb | 1.04 Å | 2020-12-16 | X-ray | 3XH | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.89% | 18.65% | 9.00% | 3-Hydroxyhippuric acid | 3-Hydroxyhippuric acid | XDS | 99.20% | 6.92 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 83.6 | - | No | - | 89.5 | 76.0 | 91.8 | - | ||||||
| 5rtc | 1.06 Å | 2020-12-16 | X-ray | EVE | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.48% | 20.08% | 9.70% | 1H-benzimidazole-2-sulfonamide | 1H-benzimidazole-2-sulfonamide | XDS | 99.80% | 6.57 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 78.9 | - | No | - | 81.2 | 71.4 | 86.6 | - | ||||||
| 5rtd | 1.04 Å | 2020-12-16 | X-ray | MHW | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.86% | 17.81% | 7.70% | 3-HYDROXYPICOLINIC ACID | 3-HYDROXYPICOLINIC ACID | XDS | 99.40% | 8.28 | SSRL (BL12-1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 82.7 | - | No | - | 92.8 | 71.4 | 89.1 | - | ||||||
| 5rte | 1.00 Å | 2020-12-16 | X-ray | 4FL | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.61% | 19.28% | 5.00% | 4-(1H-imidazol-2-yl)pyridine | 4-(1H-imidazol-2-yl)pyridine | XDS | 99.40% | 13.0 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 59.3 | - | No | - | 86.3 | 26.7 | 80.2 | - | ||||||
| 5rtf | 1.00 Å | 2020-12-16 | X-ray | ISN | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.10% | 16.59% | 3.80% | ISATIN | ISATIN | XDS | 99.30% | 18.08 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 72.4 | - | No | - | 96.0 | 38.5 | 86.8 | - | ||||||
| 5rtg | 1.01 Å | 2020-12-16 | X-ray | 3HP | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.41% | 18.00% | 7.70% | 3-HYDROXYPHENYLACETATE | 3-HYDROXYPHENYLACETATE | XDS | 99.70% | 8.19 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 77.7 | - | No | - | 92.1 | 49.1 | 94.2 | - | ||||||
| 5rth | 1.00 Å | 2020-12-16 | X-ray | 3BZ | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.71% | 17.21% | 7.40% | 3-chlorobenzoate | 3-chlorobenzoate | XDS | 99.40% | 9.32 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 73.7 | - | No | - | 94.6 | 41.0 | 89.1 | - | ||||||
| 5rti | 1.01 Å | 2020-12-16 | X-ray | 6U6 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.87% | 18.49% | 8.30% | 3-(5-chloranyl-1,3-benzothiazol-2-yl)propanoic acid | 3-(5-chloranyl-1,3-benzothiazol-2-yl)propanoic acid | XDS | 99.70% | 7.78 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 67.8 | - | No | - | 90.3 | 34.4 | 86.1 | - | ||||||
| 5rtj | 1.00 Å | 2020-12-16 | X-ray | PHB | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.19% | 17.61% | 4.30% | P-HYDROXYBENZOIC ACID | P-HYDROXYBENZOIC ACID | XDS | 99.10% | 15.78 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 70.1 | - | No | - | 93.5 | 36.4 | 86.1 | - | ||||||
| 5rtk | 1.00 Å | 2020-12-16 | X-ray | BZX | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.81% | 16.40% | 3.60% | 1,3-benzodioxol-5-ol | 1,3-benzodioxol-5-ol | XDS | 99.20% | 18.7 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 82.0 | - | No | - | 96.4 | 63.1 | 90.5 | - | ||||||
| 5rtl | 1.00 Å | 2020-12-16 | X-ray | 4J8 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.05% | 16.45% | 5.20% | 4-methylbenzenesulfonamide | 4-methylbenzenesulfonamide | XDS | 99.80% | 13.59 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 73.6 | - | No | - | 96.3 | 52.3 | 75.6 | - | ||||||
| 5rtm | 1.00 Å | 2020-12-16 | X-ray | PZA | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.25% | 16.81% | 3.90% | PYRAZINE-2-CARBOXAMIDE | PYRAZINE-2-CARBOXAMIDE | XDS | 99.20% | 18.1 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 78.8 | - | No | - | 95.5 | 55.7 | 87.7 | - | ||||||
| 5rtn | 1.00 Å | 2020-12-16 | X-ray | AQO | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.84% | 17.38% | 5.90% | 2-AMINOQUINAZOLIN-4(3H)-ONE | 2-AMINOQUINAZOLIN-4(3H)-ONE | XDS | 99.20% | 10.89 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 74.5 | - | No | - | 94.2 | 52.3 | 80.1 | - | ||||||
| 5rto | 1.00 Å | 2020-12-16 | X-ray | 4PN | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.47% | 15.84% | 6.30% | 4-PIPERIDINO-PIPERIDINE | 4-PIPERIDINO-PIPERIDINE | XDS | 99.50% | 11.43 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 77.8 | - | No | - | 97.3 | 49.1 | 89.2 | - | ||||||
| 5rtp | 1.00 Å | 2020-12-16 | X-ray | AOT | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.31% | 16.82% | 5.20% | 2-oxidanylidene-2-phenylazanyl-ethanoic acid | 2-oxidanylidene-2-phenylazanyl-ethanoic acid | XDS | 99.00% | 12.92 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 67.3 | - | No | - | 95.5 | 26.7 | 87.6 | - | ||||||
| 5rtq | 1.00 Å | 2020-12-16 | X-ray | 4JO | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.75% | 17.15% | 4.30% | 5-bromo-6-methylpyridin-2-amine | 5-bromo-6-methylpyridin-2-amine | XDS | 99.70% | 15.78 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 70.5 | - | No | - | 94.7 | 32.6 | 89.5 | - | ||||||
| 5rtr | 1.00 Å | 2020-12-16 | X-ray | SHA | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.57% | 16.82% | 5.00% | SALICYLHYDROXAMIC ACID | SALICYLHYDROXAMIC ACID | XDS | 99.20% | 13.74 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 76.7 | - | No | - | 95.5 | 52.3 | 84.6 | - | ||||||
| 5rts | 1.00 Å | 2020-12-16 | X-ray | 0LO | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.16% | 16.54% | 3.30% | 5-phenylpyridine-3-carboxylic acid | 5-phenylpyridine-3-carboxylic acid | XDS | 99.30% | 22.65 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 71.1 | - | No | - | 96.1 | 34.4 | 87.6 | - | ||||||
| 5rtt | 1.00 Å | 2020-12-16 | X-ray | NMI | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.70% | 18.20% | 4.50% | 3-(1-methyl-1H-indol-3-yl)propanoic acid | 3-(1-methyl-1H-indol-3-yl)propanoic acid | XDS | 99.20% | 14.35 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 63.2 | - | No | - | 91.4 | 27.9 | 81.8 | - | ||||||
| 5rtu | 1.00 Å | 2020-12-16 | X-ray | 1FF | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.63% | 16.27% | 4.70% | 1-methyl-5-phenyl-1H-pyrazole-4-carboxylic acid | 1-methyl-5-phenyl-1H-pyrazole-4-carboxylic acid | XDS | 98.60% | 14.57 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 81.7 | - | No | - | 96.6 | 63.1 | 89.2 | - | ||||||
| 5rtv | 1.00 Å | 2020-12-16 | X-ray | PF0 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.09% | 17.23% | 4.80% | 3-hydroxy-2-methylbenzoic acid | 3-hydroxy-2-methylbenzoic acid | XDS | 99.00% | 14.29 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 65.9 | - | No | - | 94.5 | 38.5 | 73.8 | - | ||||||
| 5rtw | 1.00 Å | 2020-12-16 | X-ray | OHP | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.05% | 17.54% | 6.70% | (2-HYDROXYPHENYL)ACETIC ACID | (2-HYDROXYPHENYL)ACETIC ACID | XDS | 98.80% | 9.79 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 70.5 | - | No | - | 93.7 | 27.9 | 95.2 | - | ||||||
| 5rtx | 1.00 Å | 2020-12-16 | X-ray | 6OT | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.60% | 16.05% | 4.80% | 3,5-dichlorobenzene-1-sulfonamide | 3,5-dichlorobenzene-1-sulfonamide | XDS | 99.40% | 14.72 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 73.7 | - | No | - | 97.0 | 38.5 | 89.1 | - | ||||||
| 5rty | 1.00 Å | 2020-12-16 | X-ray | HBD | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.45% | 18.12% | 5.20% | 4-HYDROXYBENZAMIDE | 4-HYDROXYBENZAMIDE | XDS | 99.20% | 12.64 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 76.9 | - | No | - | 91.7 | 52.3 | 89.2 | - | ||||||
| 5rtz | 1.00 Å | 2020-12-16 | X-ray | FHB | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.03% | 17.82% | 6.00% | 3-FLUORO-4-HYDROXYBENZOIC ACID | 3-FLUORO-4-HYDROXYBENZOIC ACID | XDS | 99.00% | 10.88 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 66.3 | - | No | - | 92.8 | 24.4 | 90.5 | - | ||||||
| 5ru0 | 1.00 Å | 2020-12-16 | X-ray | 2CL | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.45% | 16.64% | 5.50% | (2,6-DICHLOROPHENYL)ACETIC ACID | (2,6-DICHLOROPHENYL)ACETIC ACID | XDS | 99.00% | 12.26 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 71.5 | - | No | - | 95.9 | 43.4 | 79.7 | - | ||||||
| 5ru1 | 1.00 Å | 2020-12-16 | X-ray | DFA | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.52% | 18.04% | 4.90% | DIPHENYLACETIC ACID | DIPHENYLACETIC ACID | XDS | 99.10% | 13.4 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 67.9 | - | No | - | 92.0 | 41.0 | 78.1 | - | ||||||
| 5ru2 | 1.00 Å | 2020-12-16 | X-ray | 06Y | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.85% | 19.41% | 3.20% | 2-phenoxyethanoic acid | 2-phenoxyethanoic acid | XDS | 99.30% | 21.31 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 68.8 | - | No | - | 85.5 | 43.4 | 84.2 | - | ||||||
| 5ru3 | 1.00 Å | 2020-12-16 | X-ray | 8H8 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.62% | 16.02% | 6.60% | 2-fluoro-4-hydroxybenzonitrile | 2-fluoro-4-hydroxybenzonitrile | XDS | 99.00% | 10.93 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 82.2 | - | No | - | 97.0 | 67.2 | 86.5 | - | ||||||
| 5ru4 | 1.00 Å | 2020-12-16 | X-ray | 6V9 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.28% | 16.79% | 3.30% | 2-methyl-1,3-thiazole-5-carboxylic acid | 2-methyl-1,3-thiazole-5-carboxylic acid | XDS | 99.00% | 21.55 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 68.4 | - | No | - | 95.6 | 38.5 | 78.0 | - | ||||||
| 5ru5 | 1.00 Å | 2020-12-16 | X-ray | BXW | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.57% | 15.92% | 5.90% | 3-oxo-3,4-dihydro-2H-1,4-benzothiazine-7-carboxylic acid | 3-oxo-3,4-dihydro-2H-1,4-benzothiazine-7-carboxylic acid | XDS | 98.90% | 12.17 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 73.0 | - | No | - | 97.2 | 41.0 | 84.6 | - | ||||||
| 5ru6 | 1.00 Å | 2020-12-16 | X-ray | 2UP | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.76% | 16.45% | 7.40% | naphthalene-2-carboximidamide | naphthalene-2-carboximidamide | XDS | 99.10% | 9.16 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 78.6 | - | No | - | 96.3 | 67.2 | 74.8 | - | ||||||
| 5ru7 | 1.00 Å | 2020-12-16 | X-ray | PYD | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.72% | 16.19% | 4.10% | 2,5-DIMETHYL-PYRIMIDIN-4-YLAMINE | 2,5-DIMETHYL-PYRIMIDIN-4-YLAMINE | XDS | 99.40% | 17.49 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 75.4 | - | No | - | 96.8 | 49.1 | 83.0 | - | ||||||
| 5ru8 | 1.00 Å | 2020-12-16 | X-ray | 1SQ | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.22% | 15.58% | 3.00% | ISOQUINOLIN-1-AMINE | ISOQUINOLIN-1-AMINE | XDS | 98.90% | 23.54 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 80.7 | - | No | - | 97.7 | 55.7 | 91.8 | - | ||||||
| 5ru9 | 1.00 Å | 2020-12-16 | X-ray | 4SV | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.19% | 18.90% | 8.00% | 3-AMINOPYRIDINE-4-CARBOXYLIC ACID | 3-AMINOPYRIDINE-4-CARBOXYLIC ACID | XDS | 99.50% | 8.38 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 63.5 | - | No | - | 88.2 | 29.3 | 84.2 | - | ||||||
| 5rua | 1.00 Å | 2020-12-16 | X-ray | 3EU | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.08% | 17.45% | 5.30% | (3,5-dichlorophenyl)acetic acid | (3,5-dichlorophenyl)acetic acid | XDS | 99.00% | 12.79 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 67.9 | - | No | - | 94.0 | 38.5 | 78.6 | - | ||||||
| 5ruc | 1.00 Å | 2020-12-16 | X-ray | NCA | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.62% | 16.97% | 5.40% | NICOTINAMIDE | NICOTINAMIDE | XDS | 99.50% | 12.52 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 75.1 | - | No | - | 95.2 | 41.0 | 91.9 | - | ||||||
| 5rud | 1.00 Å | 2020-12-16 | X-ray | 2D0 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.56% | 17.12% | 6.10% | 4-chloro-1,3-benzothiazol-2-amine | 4-chloro-1,3-benzothiazol-2-amine | XDS | 99.50% | 11.46 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 73.2 | - | No | - | 94.8 | 41.0 | 87.6 | - | ||||||
| 5rue | 1.02 Å | 2020-12-16 | X-ray | BHA | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.43% | 18.30% | 7.70% | 2-HYDROXY-4-AMINOBENZOIC ACID | 2-HYDROXY-4-AMINOBENZOIC ACID | XDS | 99.80% | 8.19 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 79.0 | - | No | - | 91.0 | 59.3 | 89.2 | - | ||||||
| 5ruf | 1.00 Å | 2020-12-16 | X-ray | 54T | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.12% | 16.65% | 5.90% | 6-chloro-1,3-benzothiazol-2-amine | 6-chloro-1,3-benzothiazol-2-amine | XDS | 99.40% | 11.04 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 81.8 | - | No | - | 95.9 | 67.2 | 86.1 | - | ||||||
| 5rug | 1.00 Å | 2020-12-16 | X-ray | NOA | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.30% | 18.92% | 4.10% | NAPHTHYLOXYACETIC ACID | NAPHTHYLOXYACETIC ACID | XDS | 98.90% | 16.01 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 72.4 | - | No | - | 88.2 | 41.0 | 92.2 | - | ||||||
| 5ruh | 1.00 Å | 2020-12-16 | X-ray | KNL | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.44% | 17.15% | 5.90% | (2,6-dichlorophenoxy)acetic acid | (2,6-dichlorophenoxy)acetic acid | XDS | 98.80% | 11.92 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 63.0 | - | No | - | 94.7 | 34.4 | 71.6 | - | ||||||
| 5rui | 1.00 Å | 2020-12-16 | X-ray | 4YS | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.11% | 16.55% | 6.40% | isoquinolin-1(2H)-one | isoquinolin-1(2H)-one | XDS | 98.70% | 11.07 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 78.6 | - | No | - | 96.1 | 49.1 | 93.0 | - | ||||||
| 5ruj | 1.01 Å | 2020-12-16 | X-ray | 2SX | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.07% | 18.47% | 6.50% | (5-bromo-1H-indol-3-yl)acetic acid | (5-bromo-1H-indol-3-yl)acetic acid | XDS | 99.40% | 9.8 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 71.6 | - | No | - | 90.3 | 41.0 | 88.1 | - | ||||||
| 5ruk | 1.05 Å | 2020-12-16 | X-ray | NVU | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.42% | 18.26% | 6.10% | 2-(1,2-benzoxazol-3-yl)ethanoic acid | 2-(1,2-benzoxazol-3-yl)ethanoic acid | XDS | 99.90% | 11.4 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 72.6 | - | No | - | 91.2 | 36.4 | 94.5 | - | ||||||
| 5rul | 1.00 Å | 2020-12-16 | X-ray | 5ZE | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.67% | 17.16% | 6.30% | 4,6-dimethylpyrimidin-2-amine | 4,6-dimethylpyrimidin-2-amine | XDS | 99.50% | 11.09 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 68.7 | - | No | - | 94.7 | 36.4 | 81.8 | - | ||||||
| 5rum | 1.00 Å | 2020-12-16 | X-ray | 52F | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.14% | 17.58% | 5.10% | 3-(3-oxo-3,4-dihydroquinoxalin-2-yl)propanoic acid | 3-(3-oxo-3,4-dihydroquinoxalin-2-yl)propanoic acid | XDS | 99.30% | 13.14 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 68.5 | - | No | - | 93.6 | 32.6 | 86.1 | - | ||||||
| 5run | 1.00 Å | 2020-12-16 | X-ray | EXB | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.88% | 17.11% | 5.90% | 3-(1H-benzimidazol-1-yl)propanoic acid | 3-(1H-benzimidazol-1-yl)propanoic acid | XDS | 99.20% | 12.04 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 70.9 | - | No | - | 94.8 | 31.0 | 91.8 | - | ||||||
| 5ruo | 1.00 Å | 2020-12-16 | X-ray | W6A | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.41% | 17.86% | 4.40% | 4-chloro-1H-indole-2-carboxylic acid | 4-chloro-1H-indole-2-carboxylic acid | XDS | 99.80% | 15.21 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 69.8 | - | No | - | 92.6 | 36.4 | 86.1 | - | ||||||
| 5rup | 1.00 Å | 2020-12-16 | X-ray | 04R | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.92% | 17.36% | 4.90% | [3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl]acetic acid | [3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl]acetic acid | XDS | 99.20% | 15.05 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 57.0 | - | No | - | 94.2 | 22.4 | 72.0 | - | ||||||
| 5ruq | 1.00 Å | 2020-12-16 | X-ray | W6D | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.63% | 15.92% | 4.20% | 1H-indole-4-carboxamide | 1H-indole-4-carboxamide | XDS | 99.70% | 16.3 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 76.2 | - | No | - | 97.2 | 46.2 | 87.6 | - | ||||||
| 5rur | 1.00 Å | 2020-12-16 | X-ray | FBB | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.30% | 16.80% | 7.50% | 6-fluoro-1,3-benzothiazol-2-amine | 6-fluoro-1,3-benzothiazol-2-amine | XDS | 99.20% | 8.73 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 82.1 | - | No | - | 95.6 | 71.4 | 83.4 | - | ||||||
| 5rus | 1.00 Å | 2020-12-16 | X-ray | HSM | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.89% | 16.09% | 6.10% | HISTAMINE | HISTAMINE | XDS | 99.10% | 11.97 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 80.1 | - | No | - | 97.0 | 55.7 | 90.5 | - | ||||||
| 5rut | 1.00 Å | 2020-12-16 | X-ray | APG | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.61% | 17.83% | 7.00% | ATROLACTIC ACID (2-PHENYL-LACTIC ACID) | ATROLACTIC ACID (2-PHENYL-LACTIC ACID) | XDS | 99.60% | 9.61 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 73.1 | - | No | - | 92.7 | 38.5 | 91.8 | - | ||||||
| 5ruu | 1.01 Å | 2020-12-16 | X-ray | W6G | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.62% | 19.08% | 5.90% | N-(1,3,4-thiadiazol-2-yl)benzenesulfonamide | N-(1,3,4-thiadiazol-2-yl)benzenesulfonamide | XDS | 99.70% | 10.99 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 53.4 | - | No | - | 87.4 | 23.4 | 70.8 | - | ||||||
| 5ruv | 1.00 Å | 2020-12-16 | X-ray | W6J | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.94% | 16.28% | 3.80% | 1-(pyridin-2-yl)-1,4-diazepane | 1-(pyridin-2-yl)-1,4-diazepane | XDS | 99.30% | 18.39 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 78.4 | - | No | - | 96.6 | 52.3 | 88.8 | - | ||||||
| 5ruw | 1.00 Å | 2020-12-16 | X-ray | W6M | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.69% | 17.13% | 4.60% | 3-{[(3-methyl-1,2,4-oxadiazol-5-yl)methyl]carbamoyl}benzoic acid | 3-{[(3-methyl-1,2,4-oxadiazol-5-yl)methyl]carbamoyl}benzoic acid | XDS | 99.60% | 13.96 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 81.4 | - | No | - | 94.8 | 71.4 | 81.3 | - | ||||||
| 5rux | 1.00 Å | 2020-12-16 | X-ray | W6P | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.93% | 16.30% | 4.30% | 1,3-dihydro-2H-indol-2-one | 1,3-dihydro-2H-indol-2-one | XDS | 99.60% | 16.39 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 82.6 | - | No | - | 96.6 | 63.1 | 93.0 | - | ||||||
| 5ruy | 1.00 Å | 2020-12-16 | X-ray | XAN | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.26% | 18.93% | 3.50% | XANTHINE | XANTHINE | XDS | 98.70% | 18.88 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 67.2 | - | No | - | 88.1 | 46.2 | 75.2 | - | ||||||
| 5ruz | 1.00 Å | 2020-12-16 | X-ray | W6S | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.99% | 16.66% | 7.30% | 4-(1H-pyrazol-3-yl)piperidine | 4-(1H-pyrazol-3-yl)piperidine | XDS | 99.50% | 9.03 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 83.1 | - | No | - | 95.9 | 67.2 | 91.8 | - | ||||||
| 5rv0 | 1.00 Å | 2020-12-16 | X-ray | W6V | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.59% | 15.93% | 5.20% | N-(1,3-thiazol-2-yl)benzamide | N-(1,3-thiazol-2-yl)benzamide | XDS | 99.60% | 13.34 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 68.6 | - | No | - | 97.2 | 31.0 | 84.5 | - | ||||||
| 5rv1 | 1.00 Å | 2020-12-16 | X-ray | TFA | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.59% | 17.99% | 8.00% | trifluoroacetic acid | trifluoroacetic acid | XDS | 99.00% | 8.17 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 72.4 | - | No | - | 92.2 | 38.5 | 90.5 | - | ||||||
| 5rv2 | 1.01 Å | 2020-12-16 | X-ray | W7S | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 18.07% | 19.74% | 12.20% | N-benzylpyrazine-2-carboxamide | N-benzylpyrazine-2-carboxamide | XDS | 99.50% | 5.0 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 78.6 | - | No | - | 83.4 | 67.2 | 87.6 | - | ||||||
| 5rv3 | 1.02 Å | 2020-12-16 | X-ray | MYI | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.17% | 18.95% | 7.70% | (5-methoxy-1H-indol-3-yl)acetic acid | (5-methoxy-1H-indol-3-yl)acetic acid | XDS | 99.70% | 8.16 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 67.5 | - | No | - | 88.0 | 27.9 | 94.2 | - | ||||||
| 5rv4 | 1.00 Å | 2020-12-16 | X-ray | 4FS | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.19% | 17.65% | 8.00% | quinolin-3-amine | quinolin-3-amine | XDS | 99.80% | 8.14 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 74.3 | - | No | - | 93.4 | 38.5 | 94.2 | - | ||||||
| 5rv5 | 1.00 Å | 2020-12-16 | X-ray | JG8 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.73% | 18.25% | 3.90% | BENZOFURO[3,2-D]PYRIMIDIN-4(3H)-ONE | BENZOFURO[3,2-D]PYRIMIDIN-4(3H)-ONE | XDS | 99.70% | 16.75 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 52.7 | - | No | - | 91.2 | 24.4 | 64.8 | - | ||||||
| 5rv6 | 1.00 Å | 2020-12-16 | X-ray | 0HN | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.52% | 15.95% | 3.70% | 1,3-benzodioxole-5-carboxylic acid | 1,3-benzodioxole-5-carboxylic acid | XDS | 99.20% | 19.45 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 66.5 | - | No | - | 97.2 | 34.4 | 76.4 | - | ||||||
| 5rv7 | 1.00 Å | 2020-12-16 | X-ray | JNZ | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.64% | 16.02% | 4.70% | 1H-indazol-3-amine | 1H-indazol-3-amine | XDS | 99.40% | 14.75 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 69.4 | - | No | - | 97.0 | 32.6 | 84.6 | - | ||||||
| 5rv8 | 1.00 Å | 2020-12-16 | X-ray | 8EJ | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.08% | 17.67% | 5.70% | 6-methylpyridine-3-carboxamide | 6-methylpyridine-3-carboxamide | XDS | 99.70% | 11.45 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 82.5 | - | No | - | 93.3 | 67.2 | 91.8 | - | ||||||
| 5rv9 | 1.00 Å | 2020-12-16 | X-ray | EKZ | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.89% | 18.23% | 3.80% | 4-tert-butylbenzene-1,2-diol | 4-tert-butylbenzene-1,2-diol | XDS | 99.40% | 18.14 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 66.5 | - | No | - | 91.3 | 43.4 | 73.4 | - | ||||||
| 5rva | 1.00 Å | 2020-12-16 | X-ray | HQD | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.07% | 16.51% | 6.60% | 3-HYDROXY-2-METHYLQUINOLIN-4(1H)-ONE | 3-HYDROXY-2-METHYLQUINOLIN-4(1H)-ONE | XDS | 98.70% | 10.57 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 77.8 | - | No | - | 96.2 | 49.1 | 90.5 | - | ||||||
| 5rvb | 1.00 Å | 2020-12-16 | X-ray | 7PD | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.89% | 16.40% | 5.40% | 2-aminopteridine-4,7(3H,8H)-dione | 2-aminopteridine-4,7(3H,8H)-dione | XDS | 99.30% | 12.33 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 84.8 | - | No | - | 96.4 | 85.1 | 82.9 | - | ||||||
| 5rvc | 1.00 Å | 2020-12-16 | X-ray | W7V | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.23% | 15.57% | 3.30% | (1R,5R)-N-methyl-N-(1H-pyrazol-4-yl)bicyclo[3.1.0]hexane-1-carboxamide | (1R,5R)-N-methyl-N-(1H-pyrazol-4-yl)bicyclo[3.1.0]hexane-1-carboxamide | XDS | 99.10% | 21.79 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 82.6 | - | No | - | 97.7 | 67.2 | 87.6 | - | ||||||
| 5rvd | 1.00 Å | 2020-12-16 | X-ray | W7Y | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 18.43% | 19.81% | 4.10% | 4-[(2R)-2-cyclobutylpyrrolidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine | 4-[(2R)-2-cyclobutylpyrrolidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine | XDS | 98.90% | 16.14 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 37.8 | - | No | - | 82.9 | 15.2 | 53.6 | - | ||||||
| 5rve | 1.00 Å | 2020-12-16 | X-ray | W8A | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.75% | 16.98% | 3.90% | {[(2S)-1-oxo-1-(2-oxoimidazolidin-1-yl)propan-2-yl]sulfanyl}acetic acid | {[(2S)-1-oxo-1-(2-oxoimidazolidin-1-yl)propan-2-yl]sulfanyl}acetic acid | XDS | 99.70% | 18.76 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 74.0 | - | No | - | 95.2 | 41.0 | 89.1 | - | ||||||
| 5rvf | 1.00 Å | 2020-12-16 | X-ray | W8D | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.23% | 17.34% | 3.30% | 3-{[(2R)-oxolan-2-yl]methyl}-3H-purin-6-amine | 3-{[(2R)-oxolan-2-yl]methyl}-3H-purin-6-amine | XDS | 98.90% | 21.88 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 62.5 | - | No | - | 94.3 | 21.5 | 83.8 | - | ||||||
| 5rvg | 1.00 Å | 2020-12-16 | X-ray | W8J | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.49% | 17.94% | 3.60% | 3-{3-[(3S)-oxolan-3-yl]propyl}-3H-purin-6-amine | 3-{3-[(3S)-oxolan-3-yl]propyl}-3H-purin-6-amine | XDS | 99.50% | 18.89 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 72.1 | - | No | - | 92.3 | 43.4 | 84.7 | - | ||||||
| 5rvh | 0.98 Å | 2020-12-16 | X-ray | Q3C | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.65% | 18.38% | 6.80% | quinoline-3-carboxylic acid | quinoline-3-carboxylic acid | XDS | 99.90% | 10.18 | NSLS-II (17-ID-2) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 70.6 | - | No | - | 90.7 | 41.0 | 85.4 | - | ||||||
| 5rvi | 0.94 Å | 2020-12-16 | X-ray | CLW | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.51% | 18.09% | 6.90% | CHLORZOXAZONE | CHLORZOXAZONE | XDS | 100.00% | 10.5 | NSLS-II (17-ID-2) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 63.9 | - | No | - | 91.8 | 32.6 | 78.1 | - | ||||||
| 5rvj | 1.20 Å | 2020-12-16 | X-ray | 4JQ | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.95% | 15.83% | 3.30% | 6-amino-2H-chromen-2-one | 6-amino-2H-chromen-2-one | XDS | 85.20% | 16.4 | ALS (8.3.1) | PHENIX | 2020-09-28 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 82.1 | - | No | - | 97.3 | 63.1 | 90.3 | - | ||||||
| 5rvk | 1.46 Å | 2020-12-16 | X-ray | 2AK | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.33% | 19.46% | 5.50% | 7-bromo-5-methyl-1H-indole-2,3-dione | 7-bromo-5-methyl-1H-indole-2,3-dione | XDS | 99.20% | 11.12 | ALS (8.3.1) | PHENIX | 2020-10-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 76.6 | - | No | - | 85.2 | 71.4 | 75.5 | - | ||||||
| 5rvl | 1.36 Å | 2020-12-16 | X-ray | BVF | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.43% | 19.48% | 6.70% | 4-METHYLPYRIDIN-2-AMINE | 4-METHYLPYRIDIN-2-AMINE | XDS | 96.40% | 10.18 | ALS (8.3.1) | PHENIX | 2020-10-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 81.9 | - | No | - | 85.1 | 80.5 | 84.3 | - | ||||||
| 5rvm | 1.03 Å | 2020-12-16 | X-ray | HBD | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.28% | 15.53% | 3.50% | 4-HYDROXYBENZAMIDE | 4-HYDROXYBENZAMIDE | XDS | 78.30% | 16.79 | ALS (8.3.1) | PHENIX | 2020-10-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 73.4 | - | No | - | 97.7 | 50.2 | 75.8 | - | ||||||
| 5rvn | 1.26 Å | 2020-12-16 | X-ray | ANN | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.90% | 18.70% | 4.30% | 4-METHOXYBENZOIC ACID | 4-METHOXYBENZOIC ACID | XDS | 96.70% | 12.54 | ALS (8.3.1) | PHENIX | 2020-10-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 81.5 | - | No | - | 89.3 | 95.7 | 63.2 | - | ||||||
| 5rvo | 1.52 Å | 2020-12-16 | X-ray | AQO | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.98% | 21.19% | 7.70% | 2-AMINOQUINAZOLIN-4(3H)-ONE | 2-AMINOQUINAZOLIN-4(3H)-ONE | XDS | 97.60% | 8.34 | ALS (8.3.1) | PHENIX | 2020-10-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 80.2 | - | No | - | 72.6 | 80.5 | 90.2 | - | ||||||
| 5rvp | 1.04 Å | 2020-12-16 | X-ray | 1SQ | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.07% | 14.19% | 5.10% | ISOQUINOLIN-1-AMINE | ISOQUINOLIN-1-AMINE | XDS | 73.30% | 12.62 | SSRL (BL12-2) | PHENIX | 2020-10-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 79.5 | - | No | - | 99.0 | 95.7 | 46.3 | - | ||||||
| 5rvq | 1.08 Å | 2020-12-16 | X-ray | 4BY | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.50% | 18.33% | 9.00% | 5-methyl-1H-indole-2-carboxylic acid | 5-methyl-1H-indole-2-carboxylic acid | XDS | 79.70% | 7.66 | SSRL (BL12-2) | PHENIX | 2020-10-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 79.2 | - | No | - | 90.9 | 89.6 | 59.6 | - | ||||||
| 5rvr | 1.04 Å | 2020-12-16 | X-ray | LZ1 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.41% | 15.73% | 10.20% | 1H-indazole | 1H-indazole | XDS | 75.90% | 7.83 | SSRL (BL12-2) | PHENIX | 2020-10-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 77.7 | - | No | - | 97.5 | 80.5 | 57.5 | - | ||||||
| 5rvs | 1.52 Å | 2020-12-16 | X-ray | 0LO | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.72% | 22.26% | 8.00% | 5-phenylpyridine-3-carboxylic acid | 5-phenylpyridine-3-carboxylic acid | XDS | 97.10% | 8.66 | SSRL (BL12-2) | PHENIX | 2020-10-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 61.4 | - | No | - | 62.6 | 71.4 | 63.2 | - | ||||||
| 5rvt | 1.26 Å | 2020-12-16 | X-ray | 4BL | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.01% | 17.02% | 6.30% | 6-methyl-1H-indole-2-carboxylic acid | 6-methyl-1H-indole-2-carboxylic acid | XDS | 97.40% | 9.84 | SSRL (BL12-2) | PHENIX | 2020-10-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 80.8 | - | No | - | 95.1 | 55.7 | 94.9 | - | ||||||
| 5rvu | 1.20 Å | 2020-12-16 | X-ray | 6P3 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.61% | 18.77% | 11.70% | 6-phenylpyridine-3-carboxylic acid | 6-phenylpyridine-3-carboxylic acid | XDS | 93.50% | 6.31 | SSRL (BL12-2) | PHENIX | 2020-10-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 83.4 | - | No | - | 89.0 | 89.6 | 78.2 | - | ||||||
| 5rvv | 1.42 Å | 2020-12-16 | X-ray | WB1 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 18.15% | 21.50% | 6.30% | 6-methyl-1H-indole-3-carboxylic acid | 6-methyl-1H-indole-3-carboxylic acid | XDS | 97.30% | 10.08 | SSRL (BL12-2) | PHENIX | 2020-10-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 54.8 | - | No | - | 69.7 | 63.1 | 51.5 | - | ||||||
| 7b3o | 2.00 Å | 2020-12-16 | X-ray | - | SARS-CoV-2 | Spike | Protein-protein complex | 18.52% | 22.45% | 15.90% | - | XDS | 100.00% | 6.7 | PETRA III, DESY (P11) | PHENIX | 2020-10-02 | 34273271 | - | - | - | - | N/A | - | - | Surface glycoprotein; Light Chain of Fab Fragment; Heavy Chain of Fab Fragment | 55.6 | - | No | - | 60.7 | 36.4 | 88.7 | - | |||||||
| 7cab | 3.52 Å | 2020-12-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-12-01 | 32703908 | - | - | - | - | N/A | - | - | SARS-CoV-2 Spike glycoprotein - UNP residues 1-1208 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7cwm | 3.60 Å | 2020-12-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-06-08 | 33262452 | - | - | - | - | N/A | - | - | Spike glycoprotein; P17 heavy chain; P17 light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7cwn | 3.20 Å | 2020-12-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-08-29 | 33262452 | - | - | - | - | N/A | - | - | Spike glycoprotein; heavy chain of P17 Fab; light chain of P17 Fab; light chain of H014 Fab; heavy chain of H014 Fab | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7cwo | 3.90 Å | 2020-12-16 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | PHENIX | 2020-08-29 | 33262452 | - | - | - | - | N/A | - | - | Spike glycoprotein; heavy chain of P17 Fab; light chain of P17 Fab | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7cws | 3.40 Å | 2020-12-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-08-29 | 33262454 | - | - | - | - | N/A | - | - | Heavy Chain of FC05 Fab; Light Chain of FC05 Fab; Light Chain of H014 Fab; Heavy Chain of H014 Fab; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7cwu | 3.50 Å | 2020-12-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-08-31 | 33262454 | - | - | - | - | N/A | - | - | heavy chain of FC05 Fab; light chain of FC05 Fab; Spike glycoprotein; heavy chain of P17 Fab; light chain of P17 Fab | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7df3 | 2.70 Å | 2020-12-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-08-31 | 33277323 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7df4 | 3.80 Å | 2020-12-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-11-06 | 33277323 | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7dk3 | 6.00 Å | 2020-12-16 | Cryo-EM | VAL, ALA, TYR, ASP, PHE, SER, HIS, TRP, ILE, GLN, GLU, MET, PRO, ARG, THR, CYS, LYS, ASN | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | VALINE; HISTIDINE; PROLINE; ALANINE; TYROSINE; THREONINE; ARGININE; ASPARAGINE; CYSTEINE; SERINE; ISOLEUCINE; LEUCINE; GLUTAMIC ACID; GLUTAMINE; LYSINE; ASPARTIC ACID; METHIONINE; TRYPTOPHAN; PHENYLALANINE | VALINE; ALANINE; TYROSINE; ASPARTIC ACID; PHENYLALANINE; SERINE; HISTIDINE; TRYPTOPHAN; ISOLEUCINE; GLUTAMINE; GLUTAMIC ACID; METHIONINE; PROLINE; ARGININE; THREONINE; CYSTEINE; LYSINE; ASPARAGINE | - | - | () | 2020-11-06 | 33277323 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7kne | 3.85 Å | 2020-12-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-11-23 | 33271067 | - | - | - | - | N/A | - | - | SARS-CoV-2 spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7knh | 3.74 Å | 2020-12-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-11-04 | 33271067 | - | - | - | - | N/A | - | - | SARS-CoV-2 spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7kni | 3.91 Å | 2020-12-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-11-04 | 33271067 | - | - | - | - | N/A | - | - | SARS-CoV-2 spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7kx5 | 2.60 Å | 2020-12-16 | X-ray | X7V | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 21.01% | 27.92% | 10.90% | N-([1,1'-biphenyl]-4-yl)-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]furan-2-carboxamide | N-([1,1'-biphenyl]-4-yl)-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]furan-2-carboxamide | HKL-2000 | 97.70% | 6.4 | APS (19-ID) | REFMAC | 2020-11-04 | 34860011 | - | - | - | - | N/A | - | - | 3C-like proteinase | 27.6 | - | No | - | 14.4 | 78.5 | 37.9 | - | ||||||
| 7kyu | 1.48 Å | 2020-12-16 | X-ray | XC4 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.12% | 18.70% | 6.90% | 1-[(1H-indole-5-carbonyl)oxy]-1H-benzotriazole | 1-[(1H-indole-5-carbonyl)oxy]-1H-benzotriazole | HKL-3000 | 98.00% | 32.0 | APS (19-ID) | PHENIX | 2020-12-03 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 78.9 | - | No | - | - | 89.3 | 55.7 | 94.2 | - | |||||
| 6zlr | 3.10 Å | 2020-12-16 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 21.76% | 24.72% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | DIALS | 91.10% | 4.6 | Diamond (I24) | REFMAC | 2020-12-08 | 33381049 | - | - | - | - | N/A | - | - | Spike glycoprotein; CR3022 FAB HEAVY CHAIN; CR3022 FAB LIGHT CHAIN | 44.7 | - | No | - | 37.9 | 74.0 | 53.0 | - | |||||||
| 7b3b | 3.10 Å | 2020-12-23 | Cryo-EM | - | SARS-CoV-2 | NSP7/NSP8/NSP12/RNA | Protein-protein complex | N/A | N/A | N/A | RNA (5'-R(P*UP*GP*CP*AP*UP*CP*GP*CP*GP*UP*AP*G)-3'); DNA/RNA (5'-R(P*CP*UP*AP*CP*GP*CP*G)-D(P*(RMP))-R(P*UP*G)-3') | - | - | - | () | 2020-07-01 | 33436624 | ZN | - | - | - | N/A | - | - | SARS-CoV-2 RNA-dependent RNA polymerase (nsp12); SARS-CoV-2 nsp8; SARS-CoV-2 nsp7; DNA/RNA (5'-R(P*CP*UP*AP*CP*GP*CP*G)-D(P*(RMP))-R(P*UP*G)-3'); RNA (5'-R(P*UP*GP*CP*AP*UP*CP*GP*CP*GP*UP*AP*G)-3') | N/A | - | No | - | N/A | N/A | N/A | RNA (5'-R(P*UP*GP*CP*AP*UP*CP*GP*CP*GP*UP*AP*G)-3'); DNA/RNA (5'-R(P*CP*UP*AP*CP*GP*CP*G)-D(P*(RMP))-R(P*UP*G)-3') | - | |||||||
| 7b3c | 3.40 Å | 2020-12-23 | Cryo-EM | - | SARS-CoV-2 | NSP7/NSP8/NSP12/RNA | Protein-protein complex | N/A | N/A | N/A | RNA (5'-R(P*UP*CP*AP*CP*UP*UP*GP*CP*GP*UP*AP*G)-3'); DNA/RNA (5'-R(P*CP*UP*AP*CP*GP*CP*A)-D(P*(RMP))-R(P*GP*UP*G)-3') | - | - | - | () | 2020-11-30 | 33436624 | ZN | - | - | - | N/A | - | - | SARS-CoV-2 RNA-dependent RNA polymerase (nsp12); SARS-CoV-2 nsp8; SARS-CoV-2 nsp7; DNA/RNA (5'-R(P*CP*UP*AP*CP*GP*CP*A)-D(P*(RMP))-R(P*GP*UP*G)-3'); RNA (5'-R(P*UP*CP*AP*CP*UP*UP*GP*CP*GP*UP*AP*G)-3') | N/A | - | No | - | N/A | N/A | N/A | RNA (5'-R(P*UP*CP*AP*CP*UP*UP*GP*CP*GP*UP*AP*G)-3'); DNA/RNA (5'-R(P*CP*UP*AP*CP*GP*CP*A)-D(P*(RMP))-R(P*GP*UP*G)-3') | - | |||||||
| 7b3d | 2.80 Å | 2020-12-23 | Cryo-EM | - | SARS-CoV-2 | NSP7/NSP8/NSP12/RNA | Protein-protein complex | N/A | N/A | N/A | RNA (5'-R(P*UP*GP*CP*AP*CP*UP*GP*CP*GP*UP*AP*G)-3'); RNA (5'-R(P*CP*UP*AP*CP*GP*CP*AP*GP*UP*G)-3') | - | - | - | () | 2020-11-30 | 33436624 | ZN | - | - | - | N/A | - | - | SARS-CoV-2 RNA-dependent RNA polymerase nsp12; SARS-CoV-2 nsp8; SARS-CoV-2 nsp7; RNA (5'-R(P*CP*UP*AP*CP*GP*CP*AP*GP*UP*G)-3'); RNA (5'-R(P*UP*GP*CP*AP*CP*UP*GP*CP*GP*UP*AP*G)-3') | N/A | - | No | - | N/A | N/A | N/A | RNA (5'-R(P*CP*UP*AP*CP*GP*CP*AP*GP*UP*G)-3'); RNA (5'-R(P*UP*GP*CP*AP*CP*UP*GP*CP*GP*UP*AP*G)-3') | - | |||||||
| 7cyc | 3.21 Å | 2020-12-23 | Cryo-EM | NAG | H-CoV-229E | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-11-30 | 33420048 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7cyd | 3.55 Å | 2020-12-23 | Cryo-EM | NAG | H-CoV-229E | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-09-03 | 33420048 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7dmu | 3.20 Å | 2020-12-23 | X-ray | NAG | SARS-CoV-2 | Spike | No functional ligands | 17.88% | 19.79% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.90% | 9.38 | SPring-8 (BL44XU) | PHENIX | 2020-09-03 | 34155214 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein - receptor-binding domain | 69.9 | - | No | - | 83.2 | 85.2 | 47.1 | - | |||||||
| 7jpy | 1.60 Å | 2020-12-23 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 17.81% | 20.52% | N/A | - | iMOSFLM | 99.20% | 7.3 | ALS (5.0.2) | PHENIX | 2020-12-07 | 33283984 | - | - | - | - | N/A | - | - | 3C-like proteinase | 59.9 | - | No | - | 77.9 | 60.1 | 56.3 | - | |||||||
| 7jpz | 1.60 Å | 2020-12-23 | X-ray | GHX | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.57% | 24.09% | N/A | (phenylmethyl) N-[(2S)-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]carbamate | (phenylmethyl) N-[(2S)-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]carbamate | iMOSFLM | 97.90% | 8.8 | ALS (5.0.2) | PHENIX | 2020-08-10 | 33283984 | - | - | - | - | N/A | - | - | 3C-like proteinase | 33.6 | - | No | - | 44.1 | 45.4 | 53.8 | - | ||||||
| 7jq0 | 1.65 Å | 2020-12-23 | X-ray | VHV | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.67% | 22.71% | N/A | N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | iMOSFLM | 96.60% | 8.9 | ALS (5.0.2) | PHENIX | 2020-08-10 | 33283984 | - | - | - | - | N/A | - | - | 3C-like proteinase | 34.9 | - | No | - | 57.9 | 37.3 | 50.5 | - | ||||||
| 7jq1 | 1.65 Å | 2020-12-23 | X-ray | VHJ | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 24.74% | 29.79% | N/A | N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl]propan-2-yl}-L-phenylalaninamide | N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl]propan-2-yl}-L-phenylalaninamide | iMOSFLM | 96.50% | 7.3 | ALS (5.0.2) | PHENIX | 2020-08-10 | 33283984 | - | - | - | - | N/A | - | - | 3C-like proteinase | 7.4 | - | No | - | 7.8 | 31.1 | 40.8 | - | ||||||
| 7jq2 | 1.40 Å | 2020-12-23 | X-ray | VHM | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 15.76% | 21.39% | N/A | N-[(benzyloxy)carbonyl]-L-valyl-3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | N-[(benzyloxy)carbonyl]-L-valyl-3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | iMOSFLM | 97.00% | 10.7 | ALS (5.0.2) | PHENIX | 2020-08-10 | 33283984 | - | - | - | - | N/A | - | - | 3C-like proteinase | 43.8 | - | No | - | 70.8 | 37.4 | 54.9 | - | ||||||
| 7jq3 | 2.10 Å | 2020-12-23 | X-ray | VHP | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.66% | 25.44% | N/A | N-[(benzyloxy)carbonyl]-O-tert-butyl-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | N-[(benzyloxy)carbonyl]-O-tert-butyl-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | iMOSFLM | 99.90% | 8.8 | ROTATING ANODE () | PHENIX | 2020-08-10 | 33283984 | - | - | - | - | N/A | - | - | 3C-like proteinase | 12.7 | - | No | - | 31.3 | 25.1 | 40.0 | - | ||||||
| 7jq4 | 1.65 Å | 2020-12-23 | X-ray | XM2 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.66% | 23.82% | N/A | N-[(benzyloxy)carbonyl]-O-tert-butyl-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-phenylalaninamide | N-[(benzyloxy)carbonyl]-O-tert-butyl-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-phenylalaninamide | iMOSFLM | 99.30% | 6.1 | ALS (5.0.2) | PHENIX | 2020-08-10 | 33283984 | - | - | - | - | N/A | - | - | 3C-like proteinase | 19.4 | - | No | - | 46.6 | 26.4 | 40.0 | - | ||||||
| 7jq5 | 1.90 Å | 2020-12-23 | X-ray | NOL | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 25.97% | 32.92% | N/A | N-[(BENZYLOXY)CARBONYL]-O-(TERT-BUTYL)-L-THREONYL-3-CYCLOHEXYL-N-[(1S)-2-HYDROXY-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}ETHYL]-L-ALANINAMIDE | N-[(BENZYLOXY)CARBONYL]-O-(TERT-BUTYL)-L-THREONYL-3-CYCLOHEXYL-N-[(1S)-2-HYDROXY-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}ETHYL]-L-ALANINAMIDE | iMOSFLM | 96.60% | 5.5 | ROTATING ANODE () | PHENIX | 2020-08-10 | 33283984 | - | - | - | - | N/A | - | - | 3C-like proteinase | 12.1 | - | No | - | 4.5 | 59.9 | 30.6 | - | ||||||
| 7bbh | 2.90 Å | 2020-12-23 | Cryo-EM | NAG | Pangolin CoV | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-08-10 | 33547281 | - | - | - | - | N/A | - | - | Surface glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7l02 | 3.20 Å | 2020-12-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-11-28 | 34019795 | - | - | - | - | N/A | - | - | Spike glycoprotein; 2G12 heavy chain; 2G12 light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7l06 | 3.30 Å | 2020-12-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-12-10 | 34019795 | - | - | - | - | N/A | - | - | Spike glycoprotein; 2G12 heavy chain; 2G12 light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7l09 | 3.10 Å | 2020-12-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-12-11 | 34019795 | - | - | - | - | N/A | - | - | Spike glycoprotein; 2G12 heavy chain; 2G12 light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7l5d | 1.58 Å | 2020-12-30 | X-ray | XNJ | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.70% | 20.10% | 8.00% | N-(4-methyl-3-{[4-(pyridin-3-yl)-1,3-thiazol-2-yl]amino}phenyl)-4-[(piperazin-1-yl)methyl]benzamide | N-(4-methyl-3-{[4-(pyridin-3-yl)-1,3-thiazol-2-yl]amino}phenyl)-4-[(piperazin-1-yl)methyl]benzamide | HKL-3000 | 99.40% | 23.1 | APS (19-BM) | PHENIX | 2020-12-11 | 34285133 | - | - | - | - | N/A | - | - | 3C-like proteinase | 56.9 | - | No | - | 81.0 | 32.6 | 74.8 | - | ||||||
| 7l6r | 1.98 Å | 2020-12-30 | X-ray | BDF, MN, SAH, GLC | SARS-CoV-2 | NSP10/NSP16/RNA | Protein-protein complex | 15.04% | 16.64% | 9.50% | RNA (5'-D(*(M7G))-R(P*(A2M)P*UP*UP*AP*A)-3'); beta-D-fructopyranose; MANGANESE (II) ION; S-ADENOSYL-L-HOMOCYSTEINE; alpha-D-glucopyranose | beta-D-fructopyranose; MANGANESE (II) ION; S-ADENOSYL-L-HOMOCYSTEINE; alpha-D-glucopyranose | HKL-3000 | 100.00% | 31.2 | APS (21-ID-D) | REFMAC | 2020-12-21 | 34131072 | ZN; MN | - | - | - | N/A | - | - | 2'-O-methyltransferase; Non-structural protein 10; RNA (5'-D(*(M7G))-R(P*(A2M)P*UP*UP*AP*A)-3') | 59.9 | - | No | - | 95.9 | 12.2 | 86.2 | RNA (5'-D(*(M7G))-R(P*(A2M)P*UP*UP*AP*A)-3') | - | |||||
| 7l6t | 1.78 Å | 2021-01-06 | X-ray | BDF, FMT, SAH, GLC | SARS-CoV-2 | NSP10/NSP16/RNA | Protein-protein complex | 14.08% | 16.15% | 9.00% | RNA (5'-D(*(M7G))-R(P*(A2M)P*UP*UP*A)-3'); S-ADENOSYL-L-HOMOCYSTEINE; alpha-D-glucopyranose; beta-D-fructopyranose; FORMIC ACID | beta-D-fructopyranose; FORMIC ACID; S-ADENOSYL-L-HOMOCYSTEINE; alpha-D-glucopyranose | HKL-3000 | 96.60% | 20.8 | APS (21-ID-D) | REFMAC | 2020-12-23 | 34131072 | ZN; MG | - | - | - | N/A | - | - | 2'-O-methyltransferase; Non-structural protein 10; RNA (5'-D(*(M7G))-R(P*(A2M)P*UP*UP*A)-3') | 65.8 | - | No | - | 96.8 | 17.0 | 92.5 | RNA (5'-D(*(M7G))-R(P*(A2M)P*UP*UP*A)-3') | - | |||||
| 5s18 | 1.13 Å | 2021-01-06 | X-ray | WOY | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.53% | 21.14% | 10.40% | 6,7-dihydro-5H-pyrrolo[2,3-d]pyrimidine | 6,7-dihydro-5H-pyrrolo[2,3-d]pyrimidine | XDS | 99.90% | 5.7 | Diamond (I04-1) | BUSTER | 2020-12-23 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 63.4 | - | No | - | 72.9 | 52.1 | 76.6 | - | ||||||
| 5s1a | 1.08 Å | 2021-01-13 | X-ray | WPS | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.61% | 21.10% | 10.10% | 5-amino-3-methyl-1H-pyrazole-4-carbonitrile | 5-amino-3-methyl-1H-pyrazole-4-carbonitrile | XDS | 99.70% | 4.8 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 69.0 | - | No | - | 73.2 | 55.5 | 84.9 | - | ||||||
| 5s1c | 1.17 Å | 2021-01-13 | X-ray | WPV | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.55% | 20.16% | 8.90% | 1-(5-bromopyridin-3-yl)methanamine | 1-(5-bromopyridin-3-yl)methanamine | XDS | 99.00% | 5.4 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 81.6 | - | No | - | 80.6 | 71.3 | 96.7 | - | ||||||
| 5s1e | 1.17 Å | 2021-01-13 | X-ray | WPY | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.39% | 21.51% | 12.30% | N-(1,3-thiazol-2-yl)acetamide | N-(1,3-thiazol-2-yl)acetamide | XDS | 100.00% | 4.0 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 73.3 | - | No | - | 69.6 | 67.0 | 87.1 | - | ||||||
| 5s1g | 1.11 Å | 2021-01-13 | X-ray | WQ1 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.41% | 19.65% | 10.40% | (4-methylpyridin-3-yl)methanol | (4-methylpyridin-3-yl)methanol | XDS | 99.40% | 4.5 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 81.0 | - | No | - | 84.0 | 67.0 | 95.3 | - | ||||||
| 5s1i | 1.07 Å | 2021-01-13 | X-ray | WQ4 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.47% | 19.40% | 8.70% | 5-amino-2-methyl-1,3-oxazole-4-carbonitrile | 5-amino-2-methyl-1,3-oxazole-4-carbonitrile | XDS | 97.40% | 5.9 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 82.4 | - | No | - | 85.5 | 75.9 | 90.4 | - | ||||||
| 5s1k | 1.08 Å | 2021-01-13 | X-ray | WQ7 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.05% | 20.53% | 9.20% | 1-(2-aminoethyl)pyridin-2(1H)-one | 1-(2-aminoethyl)pyridin-2(1H)-one | XDS | 96.60% | 5.7 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 73.0 | - | No | - | 77.9 | 55.5 | 89.4 | - | ||||||
| 5s1m | 1.18 Å | 2021-01-13 | X-ray | DE5 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.85% | 20.45% | 10.90% | 2-azanyl-~{N}-(1,3-thiazol-2-yl)ethanamide | 2-azanyl-~{N}-(1,3-thiazol-2-yl)ethanamide | XDS | 99.90% | 4.5 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 74.5 | - | No | - | 78.5 | 52.1 | 96.0 | - | ||||||
| 5s1o | 1.09 Å | 2021-01-13 | X-ray | WQA | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.65% | 20.87% | 9.50% | 2H-pyrazolo[3,4-b]pyridin-5-amine | 2H-pyrazolo[3,4-b]pyridin-5-amine | XDS | 99.50% | 5.0 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 78.7 | - | No | - | 75.2 | 71.3 | 92.0 | - | ||||||
| 5s1q | 1.13 Å | 2021-01-13 | X-ray | WQG | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.43% | 20.99% | 9.90% | quinazolin-4(3H)-one | quinazolin-4(3H)-one | XDS | 99.90% | 5.5 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 71.1 | - | No | - | 74.2 | 59.2 | 84.9 | - | ||||||
| 5s1s | 1.16 Å | 2021-01-13 | X-ray | WQJ | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.02% | 20.18% | 11.70% | 7,8-dihydro-5H-pyrano[4,3-b]pyridin-3-amine | 7,8-dihydro-5H-pyrano[4,3-b]pyridin-3-amine | XDS | 99.60% | 4.0 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 80.6 | - | No | - | 80.5 | 71.3 | 93.2 | - | ||||||
| 5s1u | 1.08 Å | 2021-01-13 | X-ray | WQM | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.53% | 19.67% | 8.90% | (3S)-N-methyl-6-oxo-3,6-dihydropyridine-3-carboxamide | (3S)-N-methyl-6-oxo-3,6-dihydropyridine-3-carboxamide | XDS | 95.70% | 5.2 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 78.8 | - | No | - | 83.9 | 67.0 | 88.0 | - | ||||||
| 5s1w | 1.14 Å | 2021-01-13 | X-ray | WQV | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.78% | 20.39% | 9.30% | N-(5-bromo-2-oxo-1,2-dihydropyridin-3-yl)acetamide | N-(5-bromo-2-oxo-1,2-dihydropyridin-3-yl)acetamide | XDS | 99.90% | 5.6 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 77.6 | - | No | - | 79.0 | 62.9 | 93.2 | - | ||||||
| 5s1y | 1.09 Å | 2021-01-13 | X-ray | WQY | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.92% | 20.03% | 14.40% | 1-(quinolin-3-yl)methanamine | 1-(quinolin-3-yl)methanamine | XDS | 99.10% | 3.4 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 74.1 | - | No | - | 81.5 | 62.9 | 81.2 | - | ||||||
| 5s20 | 1.04 Å | 2021-01-13 | X-ray | WRD | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.64% | 19.61% | 7.50% | (5R)-5-amino-5,6,7,8-tetrahydronaphthalen-1-ol | (5R)-5-amino-5,6,7,8-tetrahydronaphthalen-1-ol | XDS | 96.80% | 8.0 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 82.2 | - | No | - | 84.2 | 75.9 | 90.8 | - | ||||||
| 5s22 | 1.18 Å | 2021-01-13 | X-ray | WRJ | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 18.29% | 21.77% | 13.90% | 2H-1-benzopyran-3-carboxamide | 2H-1-benzopyran-3-carboxamide | XDS | 99.90% | 3.6 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | - | - | No | - | 67.4 | - | 90.6 | - | ||||||
| 5s24 | 1.14 Å | 2021-01-13 | X-ray | WRM | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 18.47% | 22.21% | 9.70% | 2-(1H-benzimidazol-1-yl)-N-methylacetamide | 2-(1H-benzimidazol-1-yl)-N-methylacetamide | XDS | 99.90% | 5.0 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 62.7 | - | No | - | 63.0 | 46.0 | 91.1 | - | ||||||
| 5s26 | 1.12 Å | 2021-01-13 | X-ray | L46 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.34% | 20.83% | 9.30% | 4-acetyl-3-ethyl-N,5-dimethyl-1H-pyrrole-2-carboxamide | 4-acetyl-3-ethyl-N,5-dimethyl-1H-pyrrole-2-carboxamide | XDS | 99.00% | 4.8 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 75.5 | - | No | - | 75.4 | 62.9 | 90.8 | - | ||||||
| 5s27 | 1.13 Å | 2021-01-13 | X-ray | WSM | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.93% | 21.66% | 11.60% | 4-(3-aminopropyl)-2H-1,4-benzoxazin-3(4H)-one | 4-(3-aminopropyl)-2H-1,4-benzoxazin-3(4H)-one | XDS | 98.50% | 3.6 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 69.3 | - | No | - | 68.4 | 52.1 | 93.8 | - | ||||||
| 5s28 | 1.09 Å | 2021-01-13 | X-ray | WRV | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 18.10% | 21.46% | 7.70% | N-(3-fluoro-4-methylphenyl)-N'-[(2S)-1-hydroxypropan-2-yl]urea | N-(3-fluoro-4-methylphenyl)-N'-[(2S)-1-hydroxypropan-2-yl]urea | XDS | 99.50% | 7.1 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 71.1 | - | No | - | 70.2 | 62.9 | 85.3 | - | ||||||
| 5s29 | 1.30 Å | 2021-01-13 | X-ray | WRY | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.76% | 22.21% | 18.10% | 7-fluoro-N,2-dimethylquinoline-3-carboxamide | 7-fluoro-N,2-dimethylquinoline-3-carboxamide | XDS | 99.30% | 2.8 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 70.6 | - | No | - | 63.0 | 67.0 | 87.2 | - | ||||||
| 5s2a | 1.08 Å | 2021-01-13 | X-ray | WS4 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.49% | 21.06% | 8.60% | N-(4-hydroxyphenyl)-1-methyl-1H-pyrazole-4-carboxamide | N-(4-hydroxyphenyl)-1-methyl-1H-pyrazole-4-carboxamide | XDS | 99.70% | 5.8 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 66.6 | - | No | - | 73.6 | 52.1 | 82.5 | - | ||||||
| 5s2b | 1.11 Å | 2021-01-13 | X-ray | WSG | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.01% | 20.82% | 11.50% | N-(1-ethyl-1H-pyrazol-4-yl)-4-fluorobenzamide | N-(1-ethyl-1H-pyrazol-4-yl)-4-fluorobenzamide | XDS | 99.90% | 5.1 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 69.9 | - | No | - | 75.5 | 52.1 | 87.9 | - | ||||||
| 5s2c | 1.09 Å | 2021-01-13 | X-ray | WSJ | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.28% | 20.73% | 9.20% | N-(1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazol-4-yl)methanesulfonamide | N-(1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazol-4-yl)methanesulfonamide | XDS | 99.40% | 5.5 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 77.7 | - | No | - | 76.3 | 67.0 | 92.0 | - | ||||||
| 5s2d | 1.06 Å | 2021-01-13 | X-ray | U0P | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.49% | 19.31% | 7.90% | N'-cyclopropyl-N-methyl-N-[(5-methyl-1,2-oxazol-3-yl)methyl]urea | N'-cyclopropyl-N-methyl-N-[(5-methyl-1,2-oxazol-3-yl)methyl]urea | XDS | 99.10% | 7.0 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 82.2 | - | No | - | 86.1 | 71.3 | 93.4 | - | ||||||
| 5s2e | 1.12 Å | 2021-01-13 | X-ray | VZM | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.97% | 20.91% | 11.30% | N-(6-methoxypyridin-3-yl)-N'-thiophen-2-ylurea | N-(6-methoxypyridin-3-yl)-N'-thiophen-2-ylurea | XDS | 99.80% | 5.3 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 74.1 | - | No | - | 74.7 | 62.9 | 88.1 | - | ||||||
| 5s2f | 1.19 Å | 2021-01-13 | X-ray | K0G | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.00% | 21.46% | 11.60% | N-phenyl-N'-pyridin-3-ylurea | N-phenyl-N'-pyridin-3-ylurea | XDS | 99.80% | 3.5 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 77.8 | - | No | - | 70.2 | 71.3 | 94.3 | - | ||||||
| 5s2g | 1.19 Å | 2021-01-13 | X-ray | JHJ | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.25% | 21.22% | 15.30% | N-(4-methoxyphenyl)-N'-pyridin-4-ylurea | N-(4-methoxyphenyl)-N'-pyridin-4-ylurea | XDS | 100.00% | 4.5 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 67.9 | - | No | - | 72.2 | 43.3 | 95.7 | - | ||||||
| 5s2h | 1.07 Å | 2021-01-13 | X-ray | VWV | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.39% | 20.58% | 8.00% | ethyl (1,1-dioxo-1lambda~6~,4-thiazinan-4-yl)acetate | ethyl (1,1-dioxo-1lambda~6~,4-thiazinan-4-yl)acetate | XDS | 98.70% | 7.0 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 75.2 | - | No | - | 77.6 | 55.5 | 95.2 | - | ||||||
| 5s2i | 1.08 Å | 2021-01-13 | X-ray | LUY | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.40% | 20.78% | 7.90% | ~{N}-(2-phenylethyl)-1~{H}-benzimidazol-2-amine | ~{N}-(2-phenylethyl)-1~{H}-benzimidazol-2-amine | XDS | 99.50% | 7.0 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 73.3 | - | No | - | 76.0 | 59.2 | 88.5 | - | ||||||
| 5s2j | 1.11 Å | 2021-01-13 | X-ray | JGG | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.47% | 20.49% | 8.80% | N-[(4-cyanophenyl)methyl]morpholine-4-carboxamide | N-[(4-cyanophenyl)methyl]morpholine-4-carboxamide | XDS | 99.00% | 5.5 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 83.0 | - | No | - | 78.3 | 80.4 | 96.2 | - | ||||||
| 5s2k | 1.10 Å | 2021-01-13 | X-ray | VZP | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.27% | 19.25% | 8.00% | N-[(3R)-1,1-dioxo-1lambda~6~-thiolan-3-yl]-N-methyl-N'-propan-2-ylurea | N-[(3R)-1,1-dioxo-1lambda~6~-thiolan-3-yl]-N-methyl-N'-propan-2-ylurea | XDS | 99.90% | 6.4 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 83.9 | - | No | - | 86.4 | 75.9 | 97.0 | - | ||||||
| 5s2l | 1.08 Å | 2021-01-13 | X-ray | VZS | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.65% | 19.80% | 7.90% | N-(2-methoxy-5-methylphenyl)-N'-4H-1,2,4-triazol-4-ylurea | N-(2-methoxy-5-methylphenyl)-N'-4H-1,2,4-triazol-4-ylurea | XDS | 97.40% | 8.0 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 69.9 | - | No | - | 83.0 | 38.3 | 94.0 | - | ||||||
| 5s2m | 1.14 Å | 2021-01-13 | X-ray | VZY | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.54% | 19.67% | 9.30% | N-(3-methylbenzene-1-carbonyl)glycine | N-(3-methylbenzene-1-carbonyl)glycine | XDS | 99.20% | 4.5 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 80.6 | - | No | - | 83.9 | 62.9 | 98.0 | - | ||||||
| 5s2n | 1.13 Å | 2021-01-13 | X-ray | GWY | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.53% | 20.39% | 9.40% | 5-chloranyl-~{N}-methyl-~{N}-[[(3~{S})-oxolan-3-yl]methyl]pyrimidin-4-amine | 5-chloranyl-~{N}-methyl-~{N}-[[(3~{S})-oxolan-3-yl]methyl]pyrimidin-4-amine | XDS | 99.60% | 4.5 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 81.7 | - | No | - | 79.0 | 80.4 | 89.6 | - | ||||||
| 5s2o | 1.09 Å | 2021-01-13 | X-ray | NXS | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.09% | 19.54% | 5.80% | [1-(pyrimidin-2-yl)piperidin-4-yl]methanol | [1-(pyrimidin-2-yl)piperidin-4-yl]methanol | XDS | 99.30% | 9.7 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 82.2 | - | No | - | 84.7 | 71.3 | 95.0 | - | ||||||
| 5s2p | 1.03 Å | 2021-01-13 | X-ray | W04 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.92% | 19.23% | 7.70% | N~2~-methyl-N-(4-methylpyridin-2-yl)glycinamide | N~2~-methyl-N-(4-methylpyridin-2-yl)glycinamide | XDS | 98.30% | 7.4 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 82.6 | - | No | - | 86.5 | 71.3 | 95.0 | - | ||||||
| 5s2q | 1.28 Å | 2021-01-13 | X-ray | W0A | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.32% | 19.48% | 13.30% | N-[(1H-benzimidazol-2-yl)methyl]butanamide | N-[(1H-benzimidazol-2-yl)methyl]butanamide | XDS | 99.90% | 7.2 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 80.3 | - | No | - | 85.1 | 80.4 | 78.2 | - | ||||||
| 5s2r | 1.13 Å | 2021-01-13 | X-ray | K41 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.32% | 20.19% | 9.10% | 2-methyl-N-(2-methyl-2H-tetrazol-5-yl)propanamide | 2-methyl-N-(2-methyl-2H-tetrazol-5-yl)propanamide | XDS | 99.90% | 5.7 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 79.5 | - | No | - | 80.5 | 71.3 | 89.2 | - | ||||||
| 5s2s | 1.10 Å | 2021-01-13 | X-ray | GWV | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.66% | 19.43% | 9.00% | ~{N},~{N}-dimethyl-4-[(propan-2-ylamino)methyl]aniline | ~{N},~{N}-dimethyl-4-[(propan-2-ylamino)methyl]aniline | XDS | 99.90% | 6.4 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 75.4 | - | No | - | 85.4 | 48.9 | 94.5 | - | ||||||
| 5s2t | 1.11 Å | 2021-01-13 | X-ray | W0D | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.84% | 21.30% | 9.10% | N-[(1H-benzimidazol-2-yl)methyl]furan-2-carboxamide | N-[(1H-benzimidazol-2-yl)methyl]furan-2-carboxamide | XDS | 98.60% | 6.6 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 63.8 | - | No | - | 71.4 | 43.3 | 87.6 | - | ||||||
| 5s2u | 1.03 Å | 2021-01-13 | X-ray | VXD | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.39% | 19.27% | 6.70% | N-(3-chloro-2-methylphenyl)glycinamide | N-(3-chloro-2-methylphenyl)glycinamide | XDS | 98.00% | 8.5 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 82.1 | - | No | - | 86.3 | 67.0 | 97.3 | - | ||||||
| 5s2v | 1.08 Å | 2021-01-13 | X-ray | W0G | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.57% | 19.61% | 9.30% | (3R)-1-(2-fluorophenyl)-3-(methylamino)pyrrolidin-2-one | (3R)-1-(2-fluorophenyl)-3-(methylamino)pyrrolidin-2-one | XDS | 100.00% | 6.3 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 74.2 | - | No | - | 84.2 | 48.9 | 92.6 | - | ||||||
| 5s2w | 1.08 Å | 2021-01-13 | X-ray | GWP | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.80% | 20.14% | 7.80% | 2-cyclopropyl-1~{H}-imidazole-4-carboxamide | 2-cyclopropyl-1~{H}-imidazole-4-carboxamide | XDS | 99.50% | 6.4 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 77.7 | - | No | - | 80.7 | 62.9 | 91.9 | - | ||||||
| 5s2x | 1.06 Å | 2021-01-13 | X-ray | W0J | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.34% | 19.25% | 8.20% | (3R)-N-methyl-1-(pyridazin-3-yl)piperidin-3-amine | (3R)-N-methyl-1-(pyridazin-3-yl)piperidin-3-amine | XDS | 99.70% | 6.9 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 83.6 | - | No | - | 86.4 | 75.9 | 94.9 | - | ||||||
| 5s2y | 1.05 Å | 2021-01-13 | X-ray | W0M | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.94% | 20.30% | 8.30% | (2R)-2-(4-chlorophenoxy)propanamide | (2R)-2-(4-chlorophenoxy)propanamide | XDS | 98.90% | 7.3 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 72.5 | - | No | - | 79.6 | 59.2 | 82.8 | - | ||||||
| 5s2z | 1.07 Å | 2021-01-13 | X-ray | T6J | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.82% | 20.05% | 8.40% | 2-[(methylsulfonyl)methyl]-1H-benzimidazole | 2-[(methylsulfonyl)methyl]-1H-benzimidazole | XDS | 99.90% | 7.2 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 71.9 | - | No | - | 81.4 | 52.1 | 86.7 | - | ||||||
| 5s30 | 1.19 Å | 2021-01-13 | X-ray | W0P | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.53% | 20.74% | 10.00% | (2R)-2-(2-fluorophenoxy)propanoic acid | (2R)-2-(2-fluorophenoxy)propanoic acid | XDS | 99.90% | 4.3 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 75.5 | - | No | - | 76.2 | 67.0 | 85.9 | - | ||||||
| 5s31 | 1.15 Å | 2021-01-13 | X-ray | W0S | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 18.06% | 22.19% | 7.60% | 1-(3,4,5-trimethoxyphenyl)methanamine | 1-(3,4,5-trimethoxyphenyl)methanamine | XDS | 88.40% | 7.7 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 64.8 | - | No | - | 63.4 | 55.5 | 85.5 | - | ||||||
| 5s32 | 1.17 Å | 2021-01-13 | X-ray | W0V | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 20.13% | 23.64% | 53.20% | N-[(1H-benzimidazol-2-yl)methyl]-2-methylpropanamide | N-[(1H-benzimidazol-2-yl)methyl]-2-methylpropanamide | XDS | 98.40% | 3.4 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 34.3 | - | No | - | 48.5 | 11.9 | 84.3 | - | ||||||
| 5s33 | 1.06 Å | 2021-01-13 | X-ray | K2G | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.43% | 20.57% | 7.40% | 5-chloro-2-(propan-2-yl)pyrimidine-4-carboxamide | 5-chloro-2-(propan-2-yl)pyrimidine-4-carboxamide | XDS | 99.60% | 6.9 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 77.1 | - | No | - | 77.6 | 62.9 | 93.1 | - | ||||||
| 5s34 | 1.06 Å | 2021-01-13 | X-ray | GOV | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.78% | 19.35% | 7.60% | (2~{S})-1-(1,3-benzodioxol-5-ylmethylamino)propan-2-ol | (2~{S})-1-(1,3-benzodioxol-5-ylmethylamino)propan-2-ol | XDS | 97.50% | 7.3 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 83.2 | - | No | - | 85.9 | 71.3 | 98.2 | - | ||||||
| 5s35 | 1.10 Å | 2021-01-13 | X-ray | B1A | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 18.51% | 21.32% | 9.40% | ~{N}-(4-phenylazanylphenyl)ethanamide | ~{N}-(4-phenylazanylphenyl)ethanamide | XDS | 99.40% | 6.1 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 63.3 | - | No | - | 71.3 | 34.2 | 95.8 | - | ||||||
| 5s36 | 1.06 Å | 2021-01-13 | X-ray | 7ZC | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.31% | 20.93% | 7.80% | 1-(5-methoxy-1H-indol-3-yl)-N,N-dimethyl-methanamine | 1-(5-methoxy-1H-indol-3-yl)-N,N-dimethyl-methanamine | XDS | 99.20% | 8.0 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 79.8 | - | No | - | 74.6 | 71.3 | 96.0 | - | ||||||
| 5s37 | 1.22 Å | 2021-01-13 | X-ray | NZ1 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.72% | 20.53% | 11.30% | 5-methoxy-1,3-benzothiazol-2-amine | 5-methoxy-1,3-benzothiazol-2-amine | XDS | 99.90% | 6.2 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 76.8 | - | No | - | 77.9 | 67.0 | 87.9 | - | ||||||
| 5s38 | 1.07 Å | 2021-01-13 | X-ray | S7J | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.32% | 20.26% | 7.50% | 2-(trifluoromethyl)pyrimidine-5-carboxamide | 2-(trifluoromethyl)pyrimidine-5-carboxamide | XDS | 99.60% | 7.5 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 71.0 | - | No | - | 79.9 | 59.2 | 78.8 | - | ||||||
| 5s39 | 1.16 Å | 2021-01-13 | X-ray | W0Y | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.25% | 20.62% | 10.30% | N-methyl-4-sulfamoylbenzamide | N-methyl-4-sulfamoylbenzamide | XDS | 99.70% | 4.6 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 68.0 | - | No | - | 77.1 | 48.9 | 85.2 | - | ||||||
| 5s3a | 1.18 Å | 2021-01-13 | X-ray | W17 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.72% | 20.88% | 12.40% | 1-(2-hydroxyethyl)-1H-pyrazole-4-carboxamide | 1-(2-hydroxyethyl)-1H-pyrazole-4-carboxamide | XDS | 99.90% | 4.5 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 73.0 | - | No | - | 75.1 | 59.2 | 88.5 | - | ||||||
| 5s3b | 1.09 Å | 2021-01-13 | X-ray | W1A | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.16% | 19.33% | 8.40% | N-[(piperidin-4-yl)methyl]methanesulfonamide | N-[(piperidin-4-yl)methyl]methanesulfonamide | XDS | 96.60% | 5.5 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 82.1 | - | No | - | 86.0 | 75.9 | 88.5 | - | ||||||
| 5s3c | 1.19 Å | 2021-01-13 | X-ray | W1D | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.99% | 22.67% | 11.50% | (4-acetylphenoxy)acetic acid | (4-acetylphenoxy)acetic acid | XDS | 99.50% | 3.7 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 51.1 | - | No | - | 58.5 | 40.7 | 77.9 | - | ||||||
| 5s3d | 1.19 Å | 2021-01-13 | X-ray | JFP | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.89% | 21.27% | 11.30% | N-(4-methyl-1,3-thiazol-2-yl)propanamide | N-(4-methyl-1,3-thiazol-2-yl)propanamide | XDS | 99.70% | 3.6 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 52.6 | - | No | - | 71.8 | 36.0 | 72.3 | - | ||||||
| 5s3e | 1.05 Å | 2021-01-13 | X-ray | WSY | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.32% | 20.73% | 6.80% | 3-(3,5-dimethyl-1H-1,2,4-triazol-1-yl)propanoic acid | 3-(3,5-dimethyl-1H-1,2,4-triazol-1-yl)propanoic acid | XDS | 98.90% | 8.0 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 72.8 | - | No | - | 76.3 | 55.5 | 90.6 | - | ||||||
| 5s3f | 1.16 Å | 2021-01-13 | X-ray | W1J | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.53% | 19.43% | 17.60% | N-(2-propyl-2H-tetrazol-5-yl)furan-2-carboxamide | N-(2-propyl-2H-tetrazol-5-yl)furan-2-carboxamide | XDS | 99.50% | 4.2 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 80.2 | - | No | - | 85.4 | 62.9 | 95.0 | - | ||||||
| 5s3g | 1.14 Å | 2021-01-13 | X-ray | JHS | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.08% | 19.67% | 9.00% | N-[(4-phenyloxan-4-yl)methyl]acetamide | N-[(4-phenyloxan-4-yl)methyl]acetamide | XDS | 94.20% | 6.2 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 79.2 | - | No | - | 83.9 | 62.9 | 93.2 | - | ||||||
| 5s3h | 1.19 Å | 2021-01-13 | X-ray | W1M | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.61% | 21.04% | 12.80% | 3-[(1-methyl-1H-pyrazole-3-carbonyl)amino]benzoic acid | 3-[(1-methyl-1H-pyrazole-3-carbonyl)amino]benzoic acid | XDS | 99.90% | 3.4 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 77.2 | - | No | - | 73.7 | 71.3 | 88.9 | - | ||||||
| 5s3i | 1.17 Å | 2021-01-13 | X-ray | W1P | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.72% | 20.07% | 11.80% | 5-methyl-2-phenyl-2,4-dihydro-3H-pyrazol-3-one | 5-methyl-2-phenyl-2,4-dihydro-3H-pyrazol-3-one | XDS | 99.90% | 5.6 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 71.5 | - | No | - | 81.3 | 45.9 | 91.9 | - | ||||||
| 5s3j | 1.09 Å | 2021-01-13 | X-ray | W1S | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.42% | 19.67% | 8.90% | (8S)-5,6,7,8-tetrahydroimidazo[1,2-a]pyridine-8-carboxamide | (8S)-5,6,7,8-tetrahydroimidazo[1,2-a]pyridine-8-carboxamide | XDS | 95.90% | 4.5 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 81.5 | - | No | - | 83.9 | 67.0 | 97.5 | - | ||||||
| 5s3k | 1.17 Å | 2021-01-13 | X-ray | RZS, NHE | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.92% | 19.62% | 11.40% | 6-(ethylamino)pyridine-3-carbonitrile; 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID | 6-(ethylamino)pyridine-3-carbonitrile; 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID | XDS | 100.00% | 6.5 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 80.8 | - | No | - | 84.2 | 67.0 | 94.3 | - | ||||||
| 5s3l | 1.09 Å | 2021-01-13 | X-ray | JH4 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.04% | 19.64% | 9.90% | N-methylpyrimidin-2-amine | N-methylpyrimidin-2-amine | XDS | 99.70% | 5.4 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 82.1 | - | No | - | 84.1 | 71.3 | 95.3 | - | ||||||
| 5s3m | 1.26 Å | 2021-01-13 | X-ray | S2S | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.58% | 20.69% | 15.90% | 4-(methylsulfonylamino)benzamide | 4-(methylsulfonylamino)benzamide | XDS | 99.90% | 3.7 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 80.2 | - | No | - | 76.7 | 84.9 | 81.8 | - | ||||||
| 5s3n | 1.19 Å | 2021-01-13 | X-ray | W1V | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.34% | 20.02% | 11.10% | 2-(1,3,5-trimethyl-1H-pyrazol-4-yl)acetamide | 2-(1,3,5-trimethyl-1H-pyrazol-4-yl)acetamide | XDS | 99.80% | 4.1 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 80.5 | - | No | - | 81.5 | 67.0 | 96.2 | - | ||||||
| 5s3o | 1.19 Å | 2021-01-13 | X-ray | W1Y | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.70% | 19.55% | 14.60% | N-methyl-1-(1-phenyl-1H-pyrazol-4-yl)methanamine | N-methyl-1-(1-phenyl-1H-pyrazol-4-yl)methanamine | XDS | 100.00% | 5.7 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 78.9 | - | No | - | 84.7 | 67.0 | 87.5 | - | ||||||
| 5s3p | 1.10 Å | 2021-01-13 | X-ray | W21 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.16% | 19.37% | 7.60% | N-(cyclopentanecarbonyl)-L-alanine | N-(cyclopentanecarbonyl)-L-alanine | XDS | 100.00% | 7.9 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 80.8 | - | No | - | 85.8 | 62.9 | 96.8 | - | ||||||
| 5s3q | 1.09 Å | 2021-01-13 | X-ray | W2A | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.47% | 19.57% | 9.30% | (2R,3R)-2-methyl-1-(methylsulfonyl)piperidine-3-carbonitrile | (2R,3R)-2-methyl-1-(methylsulfonyl)piperidine-3-carbonitrile | XDS | 100.00% | 6.5 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 84.0 | - | No | - | 84.6 | 80.4 | 95.0 | - | ||||||
| 5s3r | 1.04 Å | 2021-01-13 | X-ray | W24 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.00% | 20.14% | 8.50% | (2S,3S)-N,2-dimethyl-1-(methylsulfonyl)piperidine-3-carboxamide | (2S,3S)-N,2-dimethyl-1-(methylsulfonyl)piperidine-3-carboxamide | XDS | 97.70% | 6.6 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 83.1 | - | No | - | 80.7 | 75.9 | 98.5 | - | ||||||
| 5s3s | 1.04 Å | 2021-01-13 | X-ray | W27 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.83% | 20.86% | 6.10% | 1-[(5S,8R)-6,7,8,9-tetrahydro-5H-5,8-epiminocyclohepta[b]pyridin-10-yl]ethan-1-one | 1-[(5S,8R)-6,7,8,9-tetrahydro-5H-5,8-epiminocyclohepta[b]pyridin-10-yl]ethan-1-one | XDS | 94.30% | 9.9 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 71.0 | - | No | - | 75.2 | 59.2 | 83.7 | - | ||||||
| 5s3t | 1.08 Å | 2021-01-13 | X-ray | W2G | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.03% | 18.82% | 12.30% | (1R,2S)-2-(thiophen-3-yl)cyclopentane-1-carboxylic acid | (1R,2S)-2-(thiophen-3-yl)cyclopentane-1-carboxylic acid | XDS | 96.70% | 4.6 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 80.7 | - | No | - | 88.7 | 67.0 | 89.7 | - | ||||||
| 5s3u | 1.08 Å | 2021-01-13 | X-ray | W2J | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.55% | 19.26% | 11.30% | [(3R,5R)-5-methylpiperidin-3-yl]methanol | [(3R,5R)-5-methylpiperidin-3-yl]methanol | XDS | 98.80% | 5.7 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 72.4 | - | No | - | 86.4 | 40.7 | 94.3 | - | ||||||
| 5s3v | 1.12 Å | 2021-01-13 | X-ray | W2M | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.90% | 19.47% | 11.10% | (2R)-1',4'-dihydro-2'H-spiro[pyrrolidine-2,3'-quinolin]-2'-one | (2R)-1',4'-dihydro-2'H-spiro[pyrrolidine-2,3'-quinolin]-2'-one | XDS | 99.30% | 4.0 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 84.2 | - | No | - | 85.2 | 84.9 | 91.0 | - | ||||||
| 5s3w | 0.99 Å | 2021-01-13 | X-ray | W2S | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.73% | 18.61% | 7.50% | (3R,4R)-4-(2-methylphenyl)oxolane-3-carboxylic acid | (3R,4R)-4-(2-methylphenyl)oxolane-3-carboxylic acid | XDS | 91.60% | 8.0 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 81.8 | - | No | - | 89.7 | 71.3 | 88.5 | - | ||||||
| 5s3x | 1.13 Å | 2021-01-13 | X-ray | W2V | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.69% | 19.70% | 10.10% | (3S,4S)-4-(3-methoxyphenyl)oxane-3-carboxylic acid | (3S,4S)-4-(3-methoxyphenyl)oxane-3-carboxylic acid | XDS | 99.70% | 7.6 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 80.5 | - | No | - | 83.7 | 62.9 | 98.0 | - | ||||||
| 5s3y | 1.11 Å | 2021-01-13 | X-ray | W2Y | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.02% | 19.75% | 11.60% | (2S,3S)-2-methyl-1-(methylsulfonyl)piperidine-3-carboxamide | (2S,3S)-2-methyl-1-(methylsulfonyl)piperidine-3-carboxamide | XDS | 95.40% | 4.0 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 82.4 | - | No | - | 83.4 | 75.9 | 92.6 | - | ||||||
| 5s3z | 1.31 Å | 2021-01-13 | X-ray | W34 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.30% | 18.58% | 49.10% | (3R,4S)-4-(3-methoxyphenyl)oxan-3-amine | (3R,4S)-4-(3-methoxyphenyl)oxan-3-amine | XDS | 96.40% | 5.7 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 85.6 | - | No | - | 89.9 | 89.6 | 89.6 | - | ||||||
| 5s40 | 1.19 Å | 2021-01-13 | X-ray | HHQ | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.68% | 20.43% | 14.50% | 4-iodanyl-3~{H}-pyridin-2-one | 4-iodanyl-3~{H}-pyridin-2-one | XDS | 99.40% | 3.3 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 78.7 | - | No | - | 78.6 | 67.0 | 92.9 | - | ||||||
| 5s41 | 1.19 Å | 2021-01-13 | X-ray | HGQ | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.74% | 21.26% | 16.30% | 4-bromanyl-1~{H}-pyridin-2-one | 4-bromanyl-1~{H}-pyridin-2-one | XDS | 99.90% | 3.7 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 72.7 | - | No | - | 71.9 | 62.9 | 87.4 | - | ||||||
| 5s42 | 1.09 Å | 2021-01-13 | X-ray | HH8 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.54% | 19.54% | 8.70% | 4-bromanyl-1,8-naphthyridine | 4-bromanyl-1,8-naphthyridine | XDS | 99.50% | 5.7 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 79.9 | - | No | - | 84.7 | 62.9 | 95.0 | - | ||||||
| 5s43 | 1.11 Å | 2021-01-13 | X-ray | UUJ | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.67% | 20.10% | 7.70% | 5-bromo-2-hydroxybenzonitrile | 5-bromo-2-hydroxybenzonitrile | XDS | 98.90% | 5.7 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 77.7 | - | No | - | 81.0 | 59.2 | 95.3 | - | ||||||
| 5s44 | 1.06 Å | 2021-01-13 | X-ray | W3A | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.56% | 20.09% | 8.90% | (4-bromo-1H-pyrazol-1-yl)acetic acid | (4-bromo-1H-pyrazol-1-yl)acetic acid | XDS | 96.90% | 6.3 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 66.2 | - | No | - | 81.2 | 34.2 | 91.9 | - | ||||||
| 5s45 | 1.16 Å | 2021-01-13 | X-ray | W3D | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.05% | 20.83% | 7.70% | (4-bromo-2-oxopyridin-1(2H)-yl)acetic acid | (4-bromo-2-oxopyridin-1(2H)-yl)acetic acid | XDS | 99.50% | 8.6 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 68.9 | - | No | - | 75.4 | 46.0 | 91.9 | - | ||||||
| 5s46 | 1.19 Å | 2021-01-13 | X-ray | HYN | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.16% | 20.55% | 13.50% | imidazolidine-2,4-dione | imidazolidine-2,4-dione | XDS | 99.90% | 3.4 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 77.4 | - | No | - | 77.7 | 71.3 | 85.6 | - | ||||||
| 5s47 | 1.09 Å | 2021-01-13 | X-ray | BAQ | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.65% | 19.65% | 9.40% | pyrrolidin-2-one | pyrrolidin-2-one | XDS | 98.40% | 6.4 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 85.0 | - | No | - | 84.0 | 84.9 | 96.6 | - | ||||||
| 5s48 | 1.07 Å | 2021-01-13 | X-ray | HRZ | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.59% | 19.92% | 14.70% | 1~{H}-pyridin-2-one | 1~{H}-pyridin-2-one | XDS | 96.90% | 3.1 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 81.9 | - | No | - | 82.2 | 75.9 | 91.7 | - | ||||||
| 5s49 | 1.03 Å | 2021-01-13 | X-ray | 3TR | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.90% | 19.74% | 10.80% | 3-AMINO-1,2,4-TRIAZOLE | 3-AMINO-1,2,4-TRIAZOLE | XDS | 94.40% | 6.3 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 56.7 | - | No | - | 83.4 | 24.2 | 80.4 | - | ||||||
| 5s4a | 1.08 Å | 2021-01-13 | X-ray | 4AP | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.42% | 19.22% | 8.00% | 4-AMINOPYRIDINE | 4-AMINOPYRIDINE | XDS | 98.10% | 5.6 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 82.9 | - | No | - | 86.6 | 71.3 | 96.2 | - | ||||||
| 5s4b | 1.19 Å | 2021-01-13 | X-ray | W3G | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.68% | 20.78% | 13.20% | pyridazin-3(2H)-one | pyridazin-3(2H)-one | XDS | 99.10% | 3.5 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 73.2 | - | No | - | 76.0 | 59.2 | 88.1 | - | ||||||
| 5s4c | 1.01 Å | 2021-01-13 | X-ray | W3J | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.44% | 18.28% | 7.90% | 1,4,5,6-tetrahydropyrimidin-2-amine | 1,4,5,6-tetrahydropyrimidin-2-amine | XDS | 96.20% | 7.7 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 79.3 | - | No | - | 91.1 | 52.4 | 96.9 | - | ||||||
| 5s4d | 1.22 Å | 2021-01-13 | X-ray | 2OP, LAC | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.50% | 20.60% | 10.80% | (2S)-2-HYDROXYPROPANOIC ACID; LACTIC ACID | (2S)-2-HYDROXYPROPANOIC ACID; LACTIC ACID | XDS | 100.00% | 5.5 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 76.6 | - | No | - | 77.3 | 71.3 | 83.6 | - | ||||||
| 5s4e | 1.07 Å | 2021-01-13 | X-ray | W3M | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.38% | 19.55% | 9.60% | 1H-imidazole-5-carbonitrile | 1H-imidazole-5-carbonitrile | XDS | 99.50% | 6.7 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 82.3 | - | No | - | 84.7 | 75.9 | 91.1 | - | ||||||
| 5s4f | 1.13 Å | 2021-01-13 | X-ray | W3P | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.37% | 21.28% | 11.00% | 1,8-naphthyridine | 1,8-naphthyridine | XDS | 99.90% | 4.6 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 70.8 | - | No | - | 71.8 | 59.2 | 86.6 | - | ||||||
| 5s4g | 1.17 Å | 2021-01-13 | X-ray | W3S | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 18.53% | 22.38% | 12.00% | [1,2,4]triazolo[4,3-a]pyridin-3-amine | [1,2,4]triazolo[4,3-a]pyridin-3-amine | XDS | 98.70% | 3.5 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | - | - | No | - | 61.5 | - | 85.4 | - | ||||||
| 5s4h | 1.18 Å | 2021-01-13 | X-ray | W3V | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.89% | 22.26% | 6.80% | 1-carbamoylpiperidine-4-carboxylic acid | 1-carbamoylpiperidine-4-carboxylic acid | XDS | 90.80% | 8.6 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 74.3 | - | No | - | 62.6 | 71.3 | 92.2 | - | ||||||
| 5s4i | 1.13 Å | 2021-01-13 | X-ray | W3Y | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.64% | 20.37% | 6.70% | (5S)-1-(4-chlorophenyl)-5-methylimidazolidine-2,4-dione | (5S)-1-(4-chlorophenyl)-5-methylimidazolidine-2,4-dione | XDS | 99.30% | 9.8 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 76.3 | - | No | - | 79.1 | 62.9 | 89.2 | - | ||||||
| 5s4j | 1.12 Å | 2021-01-13 | X-ray | W41 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.10% | 20.20% | 8.50% | 6-chlorotetrazolo[1,5-b]pyridazine | 6-chlorotetrazolo[1,5-b]pyridazine | XDS | 99.30% | 6.0 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 72.1 | - | No | - | 80.3 | 52.1 | 88.1 | - | ||||||
| 5s4k | 1.08 Å | 2021-01-13 | X-ray | W44 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.62% | 20.93% | 9.40% | (2S,5R,6R)-7-methyl-2,3,4,5,6,7-hexahydro-1H-2,6-methanoazocino[5,4-b]indol-5-ol | (2S,5R,6R)-7-methyl-2,3,4,5,6,7-hexahydro-1H-2,6-methanoazocino[5,4-b]indol-5-ol | XDS | 99.60% | 6.8 | Diamond (I04-1) | BUSTER | 2020-11-02 | 33853786 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 70.6 | - | No | - | 74.6 | 48.9 | 93.5 | - | ||||||
| 5s73 | 1.06 Å | 2021-01-13 | X-ray | - | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 20.09% | 22.34% | 7.70% | - | XDS | 99.60% | 6.3 | Diamond (I04-1) | REFMAC | 2020-11-02 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 | 66.8 | - | No | - | - | 61.8 | 75.9 | 70.9 | - | ||||||
| 5s74 | 0.96 Å | 2021-01-13 | X-ray | - | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.65% | 18.52% | 7.40% | - | XDS | 80.80% | 9.1 | Diamond (I04-1) | REFMAC | 2020-11-23 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 75.8 | - | No | - | - | 90.1 | 75.9 | 64.1 | - | ||||||
| 6lnn | 2.63 Å | 2021-01-13 | X-ray | EJC | MERS-CoV | Nucleocapsid | Functional ligand | 20.54% | 24.16% | 10.20% | 5-propoxy-1H-indole | 5-propoxy-1H-indole | HKL-2000 | 98.90% | 8.2 | NSRRC (TPS 05A) | PHENIX | 2020-11-23 | 35517857 | - | - | - | - | N/A | - | - | Nucleoprotein | 50.6 | - | No | - | 43.4 | 86.0 | 46.9 | - | ||||||
| 7b3e | 1.77 Å | 2021-01-13 | X-ray | MYC, NA | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.12% | 20.38% | 11.00% | SODIUM ION; 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE; SODIUM ION | XDS | 100.00% | 11.3 | ELETTRA (11.2C) | PHENIX; PHENIX | 2019-12-31 | 35287429 | - | - | - | - | N/A | - | - | Main Protease | 66.4 | - | No | - | 79.1 | 53.5 | 75.1 | - | ||||||
| 7b83 | 1.80 Å | 2021-01-13 | X-ray | PK8, IMD | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.41% | 20.67% | 10.70% | 9-oxa-7-thia-1-azonia-8$l^{2}-zincabicyclo[4.3.0]nona-1,3,5-triene; IMIDAZOLE | 9-oxa-7-thia-1-azonia-8$l^{2}-zincabicyclo[4.3.0]nona-1,3,5-triene; IMIDAZOLE | XDS | 99.40% | 8.8 | PETRA III, DESY (P11) | PHENIX | 2020-11-30 | 33811162 | - | - | - | - | N/A | - | - | ORF1ab polyprotein | 68.1 | - | No | - | 76.8 | 68.7 | 66.0 | - | ||||||
| 7bf3 | 2.00 Å | 2021-01-13 | X-ray | ADN | SARS-CoV-2 | NSP3: Macro | Functional ligand | 17.54% | 21.74% | 12.70% | ADENOSINE | ADENOSINE | XDS | 100.00% | 10.9 | SLS (X06SA) | REFMAC | 2020-12-12 | 33850605 | MG | - | - | - | N/A | - | - | NSP3 macrodomain | 77.4 | - | No | - | 67.6 | 79.2 | 87.9 | - | ||||||
| 7bf4 | 1.55 Å | 2021-01-13 | X-ray | 5GP | SARS-CoV-2 | NSP3: Macro | Functional ligand | 13.84% | 17.43% | 6.40% | GUANOSINE-5'-MONOPHOSPHATE | GUANOSINE-5'-MONOPHOSPHATE | XDS | 99.20% | 13.8 | SLS (X06DA) | REFMAC | 2020-12-31 | 33850605 | - | - | - | - | N/A | - | - | NSP3 macrodomain | 80.2 | - | No | - | 94.0 | 71.9 | 77.4 | - | ||||||
| 7bf5 | 2.05 Å | 2021-01-13 | X-ray | A2R | SARS-CoV-2 | NSP3: Macro | Functional ligand | 17.72% | 21.68% | 14.10% | [(2R,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3-HYDROXY-4-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4R,5R)-3,4,5-TRIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | [(2R,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3-HYDROXY-4-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4R,5R)-3,4,5-TRIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | XDS | 99.70% | 8.3 | SLS (X06SA) | REFMAC | 2020-12-31 | 33850605 | MG | - | - | - | N/A | - | - | NSP3 macrodomain | 78.6 | - | No | - | 68.2 | 84.4 | 85.7 | - | ||||||
| 7bf6 | 2.15 Å | 2021-01-13 | X-ray | F86 | SARS-CoV-2 | NSP3: Macro | Functional ligand | 18.03% | 22.58% | 9.00% | [(2~{R},3~{S},4~{R},5~{R})-5-(4-azanylpyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-cyano-3,4-bis(oxidanyl)oxolan-2-yl]methyl dihydrogen phosphate | [(2~{R},3~{S},4~{R},5~{R})-5-(4-azanylpyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-cyano-3,4-bis(oxidanyl)oxolan-2-yl]methyl dihydrogen phosphate | XDS | 99.10% | 10.7 | SLS (X06SA) | REFMAC | 2020-12-31 | 33850605 | - | - | - | - | N/A | - | - | NSP3 macrodomain | 76.6 | - | No | - | 59.5 | 80.7 | 92.1 | - | ||||||
| 7k0f | 1.65 Å | 2021-01-13 | X-ray | VR4, PG4 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.05% | 22.52% | 6.10% | N-{(2S,3R)-4-(benzylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-N~2~-[(benzyloxy)carbonyl]-L-leucinamide | N-{(2S,3R)-4-(benzylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-N~2~-[(benzyloxy)carbonyl]-L-leucinamide; TETRAETHYLENE GLYCOL | XDS | 98.30% | 10.7 | NSLS-II (17-ID-1) | PHENIX | 2020-12-31 | 34210738 | - | - | - | - | N/A | - | - | 3C-like proteinase | 64.7 | - | No | - | 60.0 | 63.5 | 80.7 | - | ||||||
| 7cm4 | 2.71 Å | 2021-01-13 | X-ray | NI | SARS-CoV-2 | Spike | Protein-protein complex | 21.66% | 24.18% | N/A | NICKEL (II) ION | NICKEL (II) ION | XDS | 99.78% | 12.44 | PAL/PLS (5C (4A)) | Coot; PHENIX | 2020-09-04 | 33436577 | NI | - | - | - | N/A | - | - | Spike glycoprotein; IgG heavy chain; IgG light chain | 41.1 | - | No | - | 43.3 | 38.8 | 76.0 | - | ||||||
| 7kn5 | 1.87 Å | 2021-01-20 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 17.68% | 21.33% | 8.40% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 95.10% | 29.0 | SSRL (BL12-1) | PHENIX | 2020-07-24 | 33436526 | - | - | - | - | N/A | - | - | Spike protein S1; VHH E; VHH U | 37.9 | - | No | - | 71.2 | 17.8 | 62.8 | - | ||||||
| 7kn6 | 2.55 Å | 2021-01-20 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 19.31% | 23.85% | 16.40% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 98.00% | 8.9 | SSRL (BL12-1) | PHENIX | 2020-11-04 | 33436526 | - | - | - | - | N/A | - | - | Spike protein S1; VHH V; CC12.3 Fab heavy chain; CC12.3 Fab light chain | 54.5 | - | No | - | 46.4 | 77.9 | 59.4 | - | ||||||
| 7kn7 | 2.73 Å | 2021-01-20 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 20.33% | 24.09% | 18.10% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 98.10% | 9.3 | APS (23-ID-B) | PHENIX | 2020-11-04 | 33436526 | - | - | - | - | N/A | - | - | Spike protein S1; VHH W; CC12.3 Fab heavy chain; CC12.3 Fab light chain | 51.9 | - | No | - | 44.1 | 70.1 | 64.4 | - | ||||||
| 7ksg | 3.33 Å | 2021-01-20 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-11-04 | 33436526 | - | - | - | - | N/A | - | - | Spike glycoprotein; Nanobody against SARS-CoV-2 glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lb7 | 2.00 Å | 2021-01-20 | X-ray/N-diffr. | SV6, DOD | SARS-CoV-2/SARS-CoV-Tor2 | NSP5 (3CLpro) | Functional ligand | 20.40% | 22.50% | 8.20% | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide; water | Mantid; CrysalisPro | 96.30% | 13.9 | ROTATING ANODE, SPALLATION SOURCE () | nCNS | 2020-11-22 | 33755450 | - | - | - | - | N/A | - | - | 3C-like proteinase | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7ldx | 2.23 Å | 2021-01-20 | X-ray | R9V | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 24.17% | 27.27% | 31.80% | (3-endo)-8-benzyl-8-azabicyclo[3.2.1]octan-3-ol | (3-endo)-8-benzyl-8-azabicyclo[3.2.1]octan-3-ol | XDS | 61.50% | 5.1 | LNLS SIRUS (MANACA) | PHENIX | 2021-01-07 | 34174328 | - | - | - | - | N/A | - | - | 3C-like proteinase - UNP residues 3264-3569 | 19.0 | - | No | - | 18.1 | 39.5 | 54.5 | - | ||||||
| 7c8j | 3.18 Å | 2021-01-27 | X-ray | - | SARS-CoV-2 | Spike | No functional ligands | 21.85% | 27.08% | 16.30% | - | HKL-2000 | 99.90% | 15.6 | FREE ELECTRON LASER () | PHENIX | 2020-06-01 | 33335073 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme; SARS-CoV-2 Receptor binding domain - UNP residues 333-527 | 26.1 | - | No | - | 19.1 | 62.3 | 46.1 | - | |||||||
| 7cwl | 3.80 Å | 2021-01-27 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-08-29 | 33262452 | - | - | - | - | N/A | - | - | Spike glycoprotein; Fab P17 heavy chain; Fab P17 light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7de1 | 2.00 Å | 2021-01-27 | X-ray | - | SARS-CoV-2 | Nucleocapsid | No functional ligands | 17.13% | 22.27% | 5.88% | - | HKL-3000 | 99.27% | 18.58 | ROTATING ANODE () | PHENIX | 2020-11-01 | 33511102 | - | - | - | - | N/A | - | - | Nucleoprotein - C-terminal RNA binding domain | 47.2 | - | No | - | 62.5 | 59.2 | 48.0 | - | |||||||
| 7kmg | 2.16 Å | 2021-01-27 | X-ray | - | SARS-CoV-2 | Spike | Protein-protein complex | 20.86% | 25.71% | 4.00% | - | XDS | 91.20% | 6.2 | APS (31-ID) | REFMAC | 2020-11-02 | 33820835 | - | - | - | - | N/A | - | - | LY-CoV555 Fab heavy chain; LY-CoV555 Fab light chain; Spike protein S1 - receptor-binding domain | 41.0 | - | No | - | 28.9 | 45.1 | 83.9 | - | |||||||
| 7kmh | 1.72 Å | 2021-01-27 | X-ray | NAG, PRO | SARS-CoV-2 | Spike | Protein-protein complex | 19.39% | 24.23% | 5.10% | 2-acetamido-2-deoxy-beta-D-glucopyranose; PROLINE | 2-acetamido-2-deoxy-beta-D-glucopyranose; PROLINE | XDS | 98.80% | 13.4 | APS (31-ID) | REFMAC | 2020-11-02 | 33820835 | - | - | - | - | N/A | - | - | LY-CoV488 Fab heavy chain; LY-CoV488 Fab light chain; Spike protein S1 - receptor-binding domain | 38.2 | - | No | - | 42.7 | 14.0 | 95.7 | - | ||||||
| 7kmi | 1.73 Å | 2021-01-27 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 18.59% | 21.72% | 3.30% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 98.70% | 5.9 | APS (31-ID) | REFMAC | 2020-11-02 | 33820835 | - | - | - | - | N/A | - | - | LY-CoV481 Fab heavy chain; LY-CoV481 Fab light chain; Spike protein S1 - receptor-binding domain | 82.6 | - | No | - | 67.7 | 93.4 | 91.5 | - | ||||||
| 7lcn | 3.35 Å | 2021-01-27 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-01-11 | 34242577 | - | - | - | - | N/A | - | - | Spike glycoprotein; DH1050.1 heavy chain; DH1050.1 light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ld1 | 3.40 Å | 2021-01-27 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-01-12 | 34242577 | - | - | - | - | N/A | - | - | Spike glycoprotein; DH1047 heavy chain; DH1047 light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7lgo | 2.45 Å | 2021-01-27 | X-ray | - | SARS-CoV-2 | NSP3: NABD | No functional ligands | 26.08% | 31.75% | 14.30% | - | HKL-3000 | 99.90% | 17.7 | ROTATING ANODE () | PHENIX | 2021-01-20 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 - nucleic acid binding domain (NAB) | 0.9 | - | No | - | - | 5.2 | 14.9 | 23.5 | - | ||||||
| 7bnm | 3.60 Å | 2021-02-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-01-22 | 33579792 | - | - | - | - | N/A | - | - | Surface glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7bnn | 3.50 Å | 2021-02-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-01-22 | 33579792 | - | - | - | - | N/A | - | - | Surface glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7bno | 4.20 Å | 2021-02-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-01-22 | 33579792 | - | - | - | - | N/A | - | - | Surface glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7kgj | 2.30 Å | 2021-02-03 | X-ray | - | SARS-CoV-2 | Spike | Protein-protein complex | 18.59% | 21.63% | 10.10% | - | XDS | 99.90% | 14.9 | APS (22-ID) | PHENIX; PHENIX | 2020-10-16 | 34537245 | - | - | - | - | N/A | - | - | Sb45, Sybody-45, Synthetic Nanobody; Spike glycoprotein - receptor binding domain | 57.0 | - | No | - | 68.6 | 70.0 | 49.9 | - | |||||||
| 7kgk | 2.60 Å | 2021-02-03 | X-ray | - | SARS-CoV-2 | Spike | Protein-protein complex | 25.80% | 27.64% | 8.00% | - | XDS | 98.80% | 18.0 | APS (22-ID) | PHENIX; PHENIX | 2020-10-16 | 34537245 | - | - | - | - | N/A | - | - | Sb16, Sybody-16, Synthetic Nanobody; Spike protein S1 - receptor binding domain | 7.7 | - | No | - | 15.9 | 22.7 | 42.1 | - | |||||||
| 7klw | 2.60 Å | 2021-02-03 | X-ray | - | SARS-CoV-2 | Spike | Protein-protein complex | 20.61% | 25.48% | 9.50% | - | XDS | 98.80% | 13.1 | APS (22-ID) | PHENIX | 2020-11-01 | 34537245 | - | - | - | - | N/A | - | - | SB68, Synthetic nanobody; SB45, Synthetic Nanobody; Spike protein S1 - RBD domain | 41.2 | - | No | - | 31.1 | 91.6 | 35.6 | - | |||||||
| 7kxj | 6.40 Å | 2021-02-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-12-04 | 34329642 | - | - | - | - | N/A | - | - | Fab 15033-7 heavy chain; Fab 15033-7 light chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7kxk | 5.00 Å | 2021-02-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-12-04 | 34329642 | - | - | - | - | N/A | - | - | Fab 15033-7 heavy chain; Fab 15033-7 light chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7kzb | 2.83 Å | 2021-02-03 | X-ray | - | SARS-CoV-2 | Spike | Protein-protein complex | 25.13% | 33.61% | 14.40% | - | XDS | 99.70% | 12.4 | Australian Synchrotron (MX2) | REFMAC | 2020-12-10 | 34024246 | - | - | - | - | N/A | - | - | Fab light chain of CR3022-B6 antibody; Fab heavy chain of CR3022-B6 antibody; Spike glycoprotein; Fab light chain of CR3014-C8 antibody; Fab heavy chain of CR3014-C8 antibody | 0.6 | - | No | - | 4.2 | 22.0 | 13.8 | - | |||||||
| 7l3n | 3.27 Å | 2021-02-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2020-12-18 | 33024963 | - | - | - | - | N/A | - | - | LY-CoV555 Fab light chain; LY-CoV555 Fab heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7lfe | 2.79 Å | 2021-02-03 | X-ray | XWS | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.50% | 25.27% | 19.80% | (2R,4R)-1-phenylhexahydropyrimidine-2,4-diol | (2R,4R)-1-phenylhexahydropyrimidine-2,4-diol | XDS | 92.10% | 9.1 | LNLS SIRUS (MANACA) | PHENIX | 2021-01-16 | 34174328 | - | - | - | - | N/A | - | - | 3C-like proteinase | 36.1 | - | No | - | 32.9 | 57.7 | 57.7 | - | ||||||
| 7lfp | 2.20 Å | 2021-02-03 | X-ray | XY4 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 21.74% | 26.10% | 14.40% | N-phenyl-N'-propan-2-ylurea | N-phenyl-N'-propan-2-ylurea | XDS | 96.80% | 6.6 | LNLS SIRUS (MANACA) | PHENIX | 2021-01-18 | 34174328 | - | - | - | - | N/A | - | - | 3C-like proteinase | 22.4 | - | No | - | 25.8 | 48.5 | 45.7 | - | ||||||
| 7b17 | 4.01 Å | 2021-02-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-11-23 | 33436526 | - | - | - | - | N/A | - | - | SARS-CoV-2 neutralizing biparatopic nanobody VE,nanobody E from Lama glama; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7klg | 3.20 Å | 2021-02-10 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 27.09% | 29.02% | 29.10% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.90% | 7.1 | CLSI (08B1-1) | PHENIX | 2020-10-30 | 34329642 | - | - | - | - | N/A | - | - | Spike glycoprotein; Fab 15033 light chain; Fab 15033 heavy chain | 29.4 | - | No | - | 9.8 | 64.4 | 60.3 | - | ||||||
| 7klh | 3.00 Å | 2021-02-10 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 25.61% | 28.35% | 17.80% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.90% | 11.18 | CLSI (08B1-1) | PHENIX | 2020-10-30 | 34329642 | - | - | - | - | N/A | - | - | Spike glycoprotein; Fab 15033-7 light chain; Fab 15033-7 heavy chain | 40.4 | - | No | - | 12.5 | 70.0 | 74.1 | - | ||||||
| 7kmk | 4.20 Å | 2021-02-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-11-03 | 34329642 | - | - | - | - | N/A | - | - | Fab 15033-7 heavy chain; Fab 15033-7 light chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7kml | 3.80 Å | 2021-02-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-11-03 | 34329642 | - | - | - | - | N/A | - | - | Fab 15033-7 heavy chain; Fab 15033-7 light chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ks9 | 4.75 Å | 2021-02-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-11-21 | 34587480 | - | - | - | - | N/A | - | - | Spike glycoprotein; 910-30 Fab heavy chain; 910-30 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7l0d | 2.39 Å | 2021-02-10 | X-ray | 0EN | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.12% | 26.10% | 6.87% | N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)furan-2-carboxamide | N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)furan-2-carboxamide | HKL-3000 | 87.91% | 12.33 | ROTATING ANODE () | PHENIX | 2020-12-11 | 33503819 | - | - | - | - | N/A | - | - | 3C-like proteinase | 22.3 | - | No | - | 25.8 | 48.7 | 45.2 | - | ||||||
| 7l1f | 3.89 Å | 2021-02-10 | Cryo-EM | - | SARS-CoV-2 | NSP7/NSP8/NSP12 | Protein-protein complex | N/A | N/A | N/A | RNA (5'-R(P*AP*UP*UP*UP*UP*AP*AP*UP*AP*GP*CP*UP*UP*CP*UP*UP*AP*G)-3'); RNA (5'-R(P*CP*UP*AP*AP*GP*AP*AP*GP*CP*UP*AP*UP*U*(F86)*(F86)*(F86)*(F86))-3') | - | - | - | () | PHENIX | 2020-12-14 | 33631104 | - | - | - | - | N/A | - | - | RNA (5'-R(P*AP*UP*UP*UP*UP*AP*AP*UP*AP*GP*CP*UP*UP*CP*UP*UP*AP*G)-3'); RNA (5'-R(P*CP*UP*AP*AP*GP*AP*AP*GP*CP*UP*AP*UP*U*(F86)*(F86)*(F86)*(F86))-3'); Non-structural protein 7; Non-structural protein 8; RNA-directed RNA polymerase | N/A | - | No | - | N/A | N/A | N/A | RNA (5'-R(P*AP*UP*UP*UP*UP*AP*AP*UP*AP*GP*CP*UP*UP*CP*UP*UP*AP*G)-3'); RNA (5'-R(P*CP*UP*AP*AP*GP*AP*AP*GP*CP*UP*AP*UP*U*(F86)*(F86)*(F86)*(F86))-3') | - | ||||||
| 7l2c | 3.65 Å | 2021-02-10 | X-ray | PGE, CAC, CA, NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 21.63% | 27.18% | N/A | CACODYLATE ION; CALCIUM ION; 2-acetamido-2-deoxy-beta-D-glucopyranose | TRIETHYLENE GLYCOL; CACODYLATE ION; CALCIUM ION; 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 93.26% | 2.57 | APS (24-ID-C) | PHENIX | 2020-12-16 | 33789084 | CA | - | - | - | N/A | - | - | 2-51 light chain; 2-51 heavy chain; Spike glycoprotein | 52.4 | - | No | - | 18.6 | 87.1 | 74.0 | - | ||||||
| 7l5b | 3.18 Å | 2021-02-10 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 18.68% | 23.87% | 11.10% | - | XDS | 99.80% | 1.7 | APS (24-ID-C) | PHENIX | 2020-12-21 | 33794145 | - | - | - | - | N/A | - | - | 2-15 Light Chain; 2-15 Heavy chain; Spike protein S1 | 47.5 | - | No | - | 46.2 | 73.2 | 50.9 | - | |||||||
| 7bh9 | 2.90 Å | 2021-02-17 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-01-11 | 34400835 | ZN | - | - | - | N/A | - | - | Surface glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7d2z | 1.97 Å | 2021-02-17 | X-ray | FMT | SARS-CoV-2 | Spike | Protein-protein complex | 18.22% | 20.70% | 9.07% | FORMIC ACID | FORMIC ACID | XDS | 99.85% | 19.48 | SSRF (BL19U1) | PHENIX | 2020-09-17 | 33657135 | - | - | - | - | N/A | - | - | Spike protein S1 - Receptor binding domain (RBD); SR31 against SARS-CoV-2 RBD, non neutralizing | 38.6 | - | No | - | 76.5 | 16.2 | 60.6 | - | ||||||
| 7d30 | 2.10 Å | 2021-02-17 | X-ray | CD, BU2, 1PE | SARS-CoV-2 | Spike | Protein-protein complex | 19.49% | 23.59% | 14.00% | 1,3-BUTANEDIOL; PENTAETHYLENE GLYCOL | CADMIUM ION; 1,3-BUTANEDIOL; PENTAETHYLENE GLYCOL | XDS | 99.70% | 12.6 | SSRF (BL19U1) | PHENIX | 2020-09-17 | 33657135 | CD | - | - | - | N/A | - | - | Spike protein S1 - Receptor binding domain (RBD); sybody fusion of MR17-SR31 with a GS linker | 40.4 | - | No | - | 49.1 | 50.4 | 57.0 | - | ||||||
| 7dpm | 3.30 Å | 2021-02-17 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 21.74% | 26.33% | 16.60% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 100.00% | 4.7 | SSRF (BL19U1) | PHENIX | 2020-12-20 | 34313527 | - | - | - | - | N/A | - | - | Spike protein S1 - RBD domain; light chain of MW06; heavy chain of MW06 | 13.9 | - | No | - | 24.2 | 27.4 | 48.4 | - | ||||||
| 7kxb | 1.55 Å | 2021-02-17 | X-ray | BME, XB1 | SARS-CoV-2 | NSP3: Macro | Functional ligand | 16.06% | 18.77% | 12.92% | BETA-MERCAPTOETHANOL; N-{3-[(1,3-dimethyl-2,6-dioxo-2,3,6,9-tetrahydro-1H-purin-8-yl)sulfanyl]propyl}-N'-[2-(morpholin-4-yl)ethyl]thiourea | BETA-MERCAPTOETHANOL; N-{3-[(1,3-dimethyl-2,6-dioxo-2,3,6,9-tetrahydro-1H-purin-8-yl)sulfanyl]propyl}-N'-[2-(morpholin-4-yl)ethyl]thiourea | XDS | 95.98% | 10.39 | SSRL (BL12-2) | PHENIX | 2020-12-03 | 33636189 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 82.0 | - | No | - | 89.0 | 71.0 | 90.3 | - | ||||||
| 7l0n | 2.78 Å | 2021-02-17 | X-ray | PGE, NAG, PG4, PG5, NA | SARS-CoV-2 | Spike | Functional ligand | 29.04% | 32.47% | 50.60% | TRIETHYLENE GLYCOL; 2-acetamido-2-deoxy-beta-D-glucopyranose; 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE; SODIUM ION | TRIETHYLENE GLYCOL; 2-acetamido-2-deoxy-beta-D-glucopyranose; TETRAETHYLENE GLYCOL; 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE; SODIUM ION | XDS | 94.80% | 3.3 | ALS (4.2.2) | REFMAC | 2020-12-11 | 33621484 | ZN; NA | - | - | - | N/A | - | - | Spike protein S1 - Binding domain; Angiotensin-converting enzyme 2; Monoclonal antibody S304 Fab heavy chain; Monoclonal antibody S304 Fab light chain; Monoclonal antibody S309 Fab heavy chain; Monoclonal antibody S309 Fab light chain | 14.1 | - | No | - | 4.7 | 21.4 | 74.1 | - | ||||||
| 7lg7 | 2.30 Å | 2021-02-17 | X-ray | XYJ | SARS-CoV-2 | NSP3: Macro | Functional ligand | 17.85% | 20.56% | 13.71% | 3-[(1,3-dimethyl-2,6-dioxo-2,3,6,9-tetrahydro-1H-purin-8-yl)sulfanyl]-N-{[2-(morpholin-4-yl)ethyl]sulfonyl}propanamide | 3-[(1,3-dimethyl-2,6-dioxo-2,3,6,9-tetrahydro-1H-purin-8-yl)sulfanyl]-N-{[2-(morpholin-4-yl)ethyl]sulfonyl}propanamide | XDS | 91.72% | 13.49 | SSRL (BL9-2) | PHENIX | 2021-01-19 | 33636189 | - | - | - | - | N/A | - | - | Non-structural protein 3 - Macrodomain | 79.9 | - | No | - | 77.7 | 71.3 | 93.3 | - | ||||||
| 7lkr | 1.65 Å | 2021-02-17 | X-ray | PG4, Y91, Y8Y, Y5S, Y4D | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.02% | 21.92% | 9.30% | (1S,2S)-2-((S)-2-(((((1R,3r,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1R,2S)-2-((S)-2-(((((1R,3r,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1S,2S)-2-((S)-2-(((((1R,3s,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1R,2S)-2-((S)-2-(((((1R,3s,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | TETRAETHYLENE GLYCOL; (1S,2S)-2-((S)-2-(((((1R,3r,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1R,2S)-2-((S)-2-(((((1R,3r,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1S,2S)-2-((S)-2-(((((1R,3s,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1R,2S)-2-((S)-2-(((((1R,3s,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | XDS | 99.90% | 8.6 | APS (17-ID) | PHENIX | 2021-02-02 | 34213885 | - | - | - | - | N/A | - | - | 3C-like proteinase | 67.2 | - | No | - | 65.8 | 53.1 | 90.7 | - | ||||||
| 7lks | 1.70 Å | 2021-02-17 | X-ray | PG4, Y4P, Y7G | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.35% | 22.11% | 7.90% | (1R,2S)-2-((S)-2-(((((1R,2S,4S)-bicyclo[2.2.1]heptan-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1S,2S)-2-((S)-2-(((((1R,2S,4S)-bicyclo[2.2.1]heptan-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | TETRAETHYLENE GLYCOL; (1R,2S)-2-((S)-2-(((((1R,2S,4S)-bicyclo[2.2.1]heptan-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1S,2S)-2-((S)-2-(((((1R,2S,4S)-bicyclo[2.2.1]heptan-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | XDS | 98.40% | 11.4 | APS (17-ID) | PHENIX | 2021-02-02 | 34213885 | - | - | - | - | N/A | - | - | 3C-like proteinase | 77.0 | - | No | - | 64.0 | 75.8 | 93.6 | - | ||||||
| 7lkt | 1.50 Å | 2021-02-17 | X-ray | PG4, FLC, Y4V, Y7M | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.79% | 20.48% | 6.30% | CITRATE ANION; (1R,2S)-2-((S)-2-(((adamantan-1-ylmethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1S,2S)-2-((S)-2-(((adamantan-1-ylmethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | TETRAETHYLENE GLYCOL; CITRATE ANION; (1R,2S)-2-((S)-2-(((adamantan-1-ylmethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1S,2S)-2-((S)-2-(((adamantan-1-ylmethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | XDS | 99.90% | 10.5 | APS (17-ID) | PHENIX | 2021-02-02 | 34213885 | - | - | - | - | N/A | - | - | 3C-like proteinase | 73.2 | - | No | - | 78.3 | 61.5 | 83.7 | - | ||||||
| 7lku | 1.65 Å | 2021-02-17 | X-ray | Y91, Y8Y, Y5S, Y4D | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.75% | 21.07% | 5.40% | (1S,2S)-2-((S)-2-(((((1R,3r,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1R,2S)-2-((S)-2-(((((1R,3r,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1S,2S)-2-((S)-2-(((((1R,3s,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1R,2S)-2-((S)-2-(((((1R,3s,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | (1S,2S)-2-((S)-2-(((((1R,3r,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1R,2S)-2-((S)-2-(((((1R,3r,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1S,2S)-2-((S)-2-(((((1R,3s,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1R,2S)-2-((S)-2-(((((1R,3s,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | XDS | 99.90% | 9.8 | APS (17-ID) | PHENIX | 2021-02-02 | 34213885 | - | - | - | - | N/A | - | - | 3C-like proteinase | 81.8 | - | No | - | 73.5 | 86.0 | 89.9 | - | ||||||
| 7lkv | 1.55 Å | 2021-02-17 | X-ray | Y64, Y4J | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.64% | 20.84% | 7.20% | (1R,2S)-2-((S)-2-(((((1R,3R,5S)-bicyclo[3.3.1]non-6-en-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | (1S,2S)-2-((S)-2-(((((1R,3R,5S)-bicyclo[3.3.1]non-6-en-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1R,2S)-2-((S)-2-(((((1R,3R,5S)-bicyclo[3.3.1]non-6-en-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | XDS | 99.40% | 8.9 | NSLS-II (17-ID-1) | PHENIX | 2021-02-02 | 34213885 | - | - | - | - | N/A | - | - | 3C-like proteinase | 75.6 | - | No | - | 75.4 | 59.3 | 94.7 | - | ||||||
| 7lkw | 1.70 Å | 2021-02-17 | X-ray | PO4, Y8V, Y8S | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.60% | 22.70% | 3.90% | PHOSPHATE ION; (1S,2S)-2-((S)-2-(((((1R,3S,5S)-bicyclo[3.3.1]non-6-en-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1R,2S)-2-((S)-2-(((((1R,3S,5S)-bicyclo[3.3.1]non-6-en-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | PHOSPHATE ION; (1S,2S)-2-((S)-2-(((((1R,3S,5S)-bicyclo[3.3.1]non-6-en-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1R,2S)-2-((S)-2-(((((1R,3S,5S)-bicyclo[3.3.1]non-6-en-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | XDS | 99.50% | 15.0 | APS (17-ID) | PHENIX | 2021-02-02 | 34213885 | - | - | - | - | N/A | - | - | 3C-like proteinase | 75.1 | - | No | - | 58.0 | 83.2 | 87.1 | - | ||||||
| 7lkx | 1.60 Å | 2021-02-17 | X-ray | PG4, Y71, Y51 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.40% | 21.23% | 7.10% | (1S,2S)-2-((S)-2-(((((1S,2S,4S)-bicyclo[2.2.1]hept-5-en-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1R,2S)-2-((S)-2-(((((1S,2S,4S)-bicyclo[2.2.1]hept-5-en-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | TETRAETHYLENE GLYCOL; (1S,2S)-2-((S)-2-(((((1S,2S,4S)-bicyclo[2.2.1]hept-5-en-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1R,2S)-2-((S)-2-(((((1S,2S,4S)-bicyclo[2.2.1]hept-5-en-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | XDS | 97.90% | 9.1 | APS (17-ID) | PHENIX | 2021-02-02 | 34213885 | - | - | - | - | N/A | - | - | 3C-like proteinase | 66.3 | - | No | - | 72.1 | 47.8 | 87.7 | - | ||||||
| 6lz6 | 2.65 Å | 2021-02-24 | X-ray | EY6 | MERS-CoV | Nucleocapsid | Functional ligand | 21.04% | 25.80% | 8.80% | 5-(2-fluoranylethoxy)-1H-indole | 5-(2-fluoranylethoxy)-1H-indole | HKL-2000 | 98.70% | 9.4 | NSRRC (TPS 05A) | PHENIX | 2020-02-18 | 35517857 | - | - | - | - | N/A | - | - | Nucleoprotein | 30.9 | - | No | - | 28.1 | 76.6 | 33.1 | - | ||||||
| 6lz8 | 2.59 Å | 2021-02-24 | X-ray | EY9 | MERS-CoV | Nucleocapsid | Functional ligand | 23.69% | 28.69% | 9.50% | 5-(2-methoxyethoxy)-1H-indole | 5-(2-methoxyethoxy)-1H-indole | HKL-2000 | 97.80% | 7.9 | NSRRC (TPS 05A) | PHENIX | 2020-02-18 | 35517857 | - | - | - | - | N/A | - | - | Nucleoprotein | 7.6 | - | No | - | 11.2 | 41.6 | 27.5 | - | ||||||
| 7cac | 3.55 Å | 2021-02-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-06-08 | 32703908 | - | - | - | - | N/A | - | - | Heavy chain of H014 Fab; Light chain of H014 Fab; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7l7f | 3.24 Å | 2021-02-24 | Cryo-EM | - | HIV-1/SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2020-12-28 | 33524990 | - | - | - | - | N/A | - | - | Spike glycoprotein, Envelope glycoprotein fusion; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7l7k | 3.29 Å | 2021-02-24 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | PHENIX | 2020-12-28 | 33524990 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lbn | 1.76 Å | 2021-02-24 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 15.98% | 18.55% | 3.80% | Calpain I Inhibitor | - | HKL-2000 | 97.00% | 38.4 | CHESS (A1) | REFMAC | 2021-01-08 | 35217718 | - | - | - | - | N/A | - | - | Calpain I Inhibitor; 3C-like proteinase | 70.6 | - | No | - | 90.0 | 29.2 | 97.7 | - | ||||||
| 7lbr | 2.20 Å | 2021-02-24 | X-ray | XT7 | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 19.59% | 24.06% | 13.10% | 5-[(azetidin-3-yl)amino]-N-[(1R)-1-{3-[5-({[(1S,3R)-3-hydroxycyclopentyl]amino}methyl)thiophen-2-yl]phenyl}ethyl]-2-methylbenzamide | 5-[(azetidin-3-yl)amino]-N-[(1R)-1-{3-[5-({[(1S,3R)-3-hydroxycyclopentyl]amino}methyl)thiophen-2-yl]phenyl}ethyl]-2-methylbenzamide | Aimless | 98.50% | 6.5 | APS (21-ID-D) | REFMAC | 2021-01-08 | 33594371 | ZN | - | - | - | N/A | - | - | Non-structural protein 3 - residues 1564-1878 | 44.2 | - | No | - | 44.3 | 33.3 | 86.4 | - | ||||||
| 7lbs | 2.80 Å | 2021-02-24 | X-ray | BO3, XR8 | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 20.16% | 26.01% | 20.00% | BORIC ACID; 5-[(azetidin-3-yl)amino]-2-methyl-N-[(1R)-1-(3-{5-[(pyrrolidin-1-yl)methyl]thiophen-2-yl}phenyl)ethyl]benzamide | BORIC ACID; 5-[(azetidin-3-yl)amino]-2-methyl-N-[(1R)-1-(3-{5-[(pyrrolidin-1-yl)methyl]thiophen-2-yl}phenyl)ethyl]benzamide | XDS | 79.60% | 6.8 | APS (21-ID-F) | REFMAC | 2021-01-08 | 33594371 | ZN | - | - | - | N/A | - | - | Non-structural protein 3 - residues 1564-1878 | 25.5 | - | No | - | 26.4 | 67.5 | 32.6 | - | ||||||
| 7llf | 2.30 Å | 2021-02-24 | X-ray | Y54, BO3 | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 21.74% | 25.98% | 16.40% | 5-[(azetidin-3-yl)amino]-N-[(1R)-1-{3-[5-({[(1R,3S)-3-hydroxycyclopentyl]amino}methyl)thiophen-2-yl]phenyl}ethyl]-2-methylbenzamide; BORIC ACID | 5-[(azetidin-3-yl)amino]-N-[(1R)-1-{3-[5-({[(1R,3S)-3-hydroxycyclopentyl]amino}methyl)thiophen-2-yl]phenyl}ethyl]-2-methylbenzamide; BORIC ACID | Aimless | 95.10% | 5.0 | APS (21-ID-D) | REFMAC | 2021-02-03 | 33594371 | ZN | - | - | - | N/A | - | - | Non-structural protein 3 | 39.1 | - | No | - | 26.8 | 63.3 | 64.0 | - | ||||||
| 7llz | 2.90 Å | 2021-02-24 | X-ray | Y61 | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 19.59% | 24.27% | 17.90% | N-[(1R)-1-(3-{5-[(acetylamino)methyl]thiophen-2-yl}phenyl)ethyl]-5-[(azetidin-3-yl)amino]-2-methylbenzamide | N-[(1R)-1-(3-{5-[(acetylamino)methyl]thiophen-2-yl}phenyl)ethyl]-5-[(azetidin-3-yl)amino]-2-methylbenzamide | xia2 | 99.90% | 8.1 | APS, APS (21-ID-D, 21-ID-G) | REFMAC | 2021-02-04 | 33594371 | ZN | - | - | - | N/A | - | - | Non-structural protein 3 | 65.1 | - | No | - | 42.4 | 79.3 | 83.4 | - | ||||||
| 7los | 2.90 Å | 2021-02-24 | X-ray | Y97 | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 20.94% | 29.29% | N/A | 5-(azetidin-3-ylamino)-2-methyl-~{N}-[(1~{R})-1-[3-[5-[[[(3~{R})-oxolan-3-yl]amino]methyl]thiophen-2-yl]phenyl]ethyl]benzamide | 5-(azetidin-3-ylamino)-2-methyl-~{N}-[(1~{R})-1-[3-[5-[[[(3~{R})-oxolan-3-yl]amino]methyl]thiophen-2-yl]phenyl]ethyl]benzamide | XDS | 98.10% | 4.7 | APS (21-ID-D) | REFMAC | 2021-02-10 | 33594371 | ZN | - | - | - | N/A | - | - | Non-structural protein 3 | 15.8 | - | No | - | 9.1 | 53.5 | 41.9 | - | ||||||
| 7nfv | 1.42 Å | 2021-02-24 | X-ray | PO4 | SARS-CoV-2 | NSP3: PLpro | No functional ligands | 15.38% | 17.14% | N/A | PHOSPHATE ION | PHOSPHATE ION | XDS | 100.00% | 28.3 | PETRA III, DESY (P11) | REFMAC | 2021-02-07 | 35953531 | ZN | - | - | - | N/A | - | - | 2'-O-methyltransferase | 70.1 | - | No | - | 94.8 | 46.8 | 74.3 | - | ||||||
| 7aqe | 1.39 Å | 2021-03-03 | X-ray | RV5 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.66% | 22.31% | N/A | N-1,2,3-Benzothiadiazol-6-yl-N'-[2-oxo-2-(1-piperidinyl)ethyl]urea also called unc-2327 | DIALS | 98.93% | 8.11 | PETRA III, DESY (P11) | REFMAC; PHENIX | 2020-10-21 | 33811162 | - | - | - | - | N/A | - | - | Structure of SARS-CoV-2 Main Protease bound to UNC-2327 | 51.6 | - | No | - | 62.0 | 37.5 | 78.7 | - | |||||||
| 7bb2 | 1.60 Å | 2021-03-03 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 16.42% | 18.86% | 9.60% | SODIUM ION | SODIUM ION | XDS | 100.00% | 15.2 | ELETTRA (11.2C) | PHENIX; PHENIX | 2020-12-16 | 34769210 | - | - | - | - | N/A | - | - | Main Protease | 78.9 | - | No | - | 88.5 | 64.1 | 86.4 | - | ||||||
| 7be7 | 1.68 Å | 2021-03-03 | X-ray | NA, ALD | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.66% | 20.13% | 9.80% | SODIUM ION; N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | SODIUM ION; N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | XDS | 99.90% | 11.9 | ELETTRA (11.2C) | PHENIX; PHENIX | 2020-12-22 | 34769210 | - | - | - | - | N/A | - | - | Main Protease | 68.9 | - | No | - | 80.8 | 53.9 | 78.6 | - | ||||||
| 7beh | 2.30 Å | 2021-03-03 | X-ray | TRS | SARS-CoV-2 | Spike | Functional ligand | 21.32% | 22.96% | 10.70% | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL | xia2 | 85.70% | 14.2 | Diamond (I03) | PHENIX | 2020-12-23 | 33756110 | - | - | - | - | N/A | - | - | COVOX-316 light chain; COVOX-316 heavy chain; Spike glycoprotein | 48.6 | - | No | - | 55.5 | 40.9 | 76.1 | - | ||||||
| 7bei | 2.30 Å | 2021-03-03 | X-ray | NO3 | SARS-CoV-2 | Spike | Protein-protein complex | 18.37% | 23.19% | 19.20% | NITRATE ION | NITRATE ION | xia2 | 84.40% | 12.5 | Diamond (I03) | PHENIX | 2020-12-23 | 33756110 | - | - | - | - | N/A | - | - | Spike glycoprotein; COVOX-150 light chain; COVOX-150 heavy chain | 54.4 | - | No | - | 53.3 | 62.9 | 67.3 | - | ||||||
| 7bej | 2.42 Å | 2021-03-03 | X-ray | FMT | SARS-CoV-2 | Spike | Protein-protein complex | 18.96% | 23.14% | 25.00% | FORMIC ACID | FORMIC ACID | xia2 | 98.50% | 7.6 | Diamond (I03) | PHENIX | 2020-12-23 | 33756110 | - | - | - | - | N/A | - | - | Spike glycoprotein; COVOX-158 light chain; COVOX-158 heavy chain | 64.8 | - | No | - | 53.6 | 74.1 | 76.8 | - | ||||||
| 7bek | 2.04 Å | 2021-03-03 | X-ray | TRS | SARS-CoV-2 | Spike | Functional ligand | 19.97% | 21.95% | 11.80% | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL | xia2 | 99.90% | 13.1 | Diamond (I03) | PHENIX | 2020-12-23 | 33756110 | - | - | - | - | N/A | - | - | Spike glycoprotein; COVOX-158 light chain; COVOX-158 heavy chain | 49.8 | - | No | - | 65.6 | 24.2 | 84.8 | - | ||||||
| 7bel | 2.53 Å | 2021-03-03 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 24.94% | 28.56% | 18.90% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 97.80% | 5.6 | Diamond (I03) | PHENIX | 2020-12-23 | 33756110 | - | - | - | - | N/A | - | - | COVOX-45 light chain; COVOX-88 light chain; COVOX-88 heavy chain; COVOX-45 heavy chain; Spike glycoprotein | 11.9 | - | No | - | 11.7 | 23.5 | 59.0 | - | ||||||
| 7bem | 2.52 Å | 2021-03-03 | X-ray | PRO, PO4 | SARS-CoV-2 | Spike | Protein-protein complex | 21.69% | 24.69% | 43.20% | PROLINE; PHOSPHATE ION | PROLINE; PHOSPHATE ION | xia2 | 99.90% | 9.2 | Diamond (I03) | PHENIX | 2020-12-24 | 33756110 | - | - | - | - | N/A | - | - | Spike glycoprotein; COVOX-269 Vl domain; COVOX-269 Vh domain | 29.1 | - | No | - | 38.4 | 24.2 | 71.4 | - | ||||||
| 7ben | 2.50 Å | 2021-03-03 | X-ray | BR, IMD, IOD, PG6 | SARS-CoV-2 | Spike | Protein-protein complex | 24.04% | 28.22% | 28.30% | BROMIDE ION; IMIDAZOLE; IODIDE ION; 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE | BROMIDE ION; IMIDAZOLE; IODIDE ION; 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE | xia2 | 100.00% | 7.3 | Diamond (I03) | PHENIX | 2020-12-24 | 33756110 | - | - | - | - | N/A | - | - | COVOX-75 light chain; COVOX-75 heavy chain; COVOX-253 light chain; COVOX-253 heavy chain; Spike glycoprotein | 23.6 | - | No | - | 13.0 | 43.6 | 65.8 | - | ||||||
| 7beo | 3.19 Å | 2021-03-03 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 23.41% | 27.35% | 41.20% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 100.00% | 4.0 | Diamond (I03) | PHENIX | 2020-12-24 | 33756110 | - | - | - | - | N/A | - | - | COVOX-75 light chain; COVOX-75 heavy chain; COVOX-253H55L light chain; COVOX-253H55L heavy chain; Spike glycoprotein | 21.4 | - | No | - | 17.5 | 25.0 | 75.2 | - | ||||||
| 7bep | 2.61 Å | 2021-03-03 | X-ray | GLU, IMD, PG4, PGE | SARS-CoV-2 | Spike | Protein-protein complex | 20.25% | 24.08% | 11.20% | GLUTAMIC ACID; IMIDAZOLE; alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose; TRIETHYLENE GLYCOL | GLUTAMIC ACID; IMIDAZOLE; TETRAETHYLENE GLYCOL; TRIETHYLENE GLYCOL | xia2 | 100.00% | 13.3 | Diamond (I03) | PHENIX | 2020-12-24 | 33756110 | - | - | - | - | N/A | - | - | S309 light chain; S309 heavy chain; COVOX-384 light chain; COVOX-384 heavy chain; Spike glycoprotein | 43.4 | - | No | - | 44.2 | 50.9 | 67.4 | - | ||||||
| 7bfb | 2.05 Å | 2021-03-03 | X-ray | NA, 9JT | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.38% | 19.91% | 8.60% | SODIUM ION; ~{N}-phenyl-2-selanyl-benzamide | SODIUM ION; ~{N}-phenyl-2-selanyl-benzamide | XDS | 99.80% | 12.3 | ELETTRA (11.2C) | PHENIX; PHENIX | 2021-01-02 | - | - | - | - | - | N/A | - | - | Main Protease | 70.6 | - | No | - | - | 82.3 | 50.2 | 84.6 | - | |||||
| 7bgp | 1.68 Å | 2021-03-03 | X-ray | NA, ALD | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.04% | 20.10% | 9.20% | SODIUM ION; N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | SODIUM ION; N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | XDS | 99.80% | 12.5 | ELETTRA (11.2C) | PHENIX; PHENIX | 2021-01-08 | 34769210 | - | - | - | - | N/A | - | - | Main Protease | 63.0 | - | No | - | 81.0 | 41.0 | 78.8 | - | ||||||
| 7ci3 | 2.20 Å | 2021-03-03 | X-ray | - | SARS-CoV-2 | ORF7a | No functional ligands | 22.95% | 25.98% | 10.30% | - | HKL-3000 | 99.70% | 4.7 | SSRF (BL18U1) | PHENIX | 2020-07-07 | 33615195 | - | - | - | - | N/A | - | - | Orf7a protein | 35.6 | - | No | - | 26.8 | 56.6 | 63.8 | - | |||||||
| 7cn4 | 2.93 Å | 2021-03-03 | Cryo-EM | NAG | Bat-CoV-RaTG13 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-07-30 | 33707453 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7cn8 | 2.50 Å | 2021-03-03 | Cryo-EM | EIC, NAG | Pangolin CoV | Spike | Functional ligand | N/A | N/A | N/A | LINOLEIC ACID; 2-acetamido-2-deoxy-beta-D-glucopyranose | LINOLEIC ACID; 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-07-30 | 33707453 | - | - | - | - | N/A | - | - | Glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7jst | 1.85 Å | 2021-03-03 | X-ray | PO4 | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 16.80% | 19.61% | N/A | PHOSPHATE ION | PHOSPHATE ION | XDS | 98.90% | 13.4 | APS (24-ID-C) | PHENIX | 2020-08-16 | 33795671 | - | - | - | - | N/A | - | - | 3C-like proteinase | 83.2 | - | No | - | 84.2 | 88.6 | 82.6 | - | ||||||
| 7l10 | 1.63 Å | 2021-03-03 | X-ray | XEY | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 22.43% | 25.07% | N/A | 2-[3-(3,5-dichlorophenyl)-2-oxo[2H-[1,3'-bipyridine]]-5-yl]benzonitrile | 2-[3-(3,5-dichlorophenyl)-2-oxo[2H-[1,3'-bipyridine]]-5-yl]benzonitrile | XDS | 96.60% | 7.16 | NSLS-II (17-ID-1) | PHENIX | 2020-12-13 | 33786375 | - | - | - | - | N/A | - | - | 3C-like proteinase | 19.8 | - | No | - | 34.6 | 22.8 | 56.5 | - | ||||||
| 7l11 | 1.80 Å | 2021-03-03 | X-ray | XF1 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.51% | 23.16% | 9.20% | 2-[3-(3-chloro-5-propoxyphenyl)-2-oxo[2H-[1,3'-bipyridine]]-5-yl]benzonitrile | 2-[3-(3-chloro-5-propoxyphenyl)-2-oxo[2H-[1,3'-bipyridine]]-5-yl]benzonitrile | XDS | 99.20% | 16.17 | APS (24-ID-E) | PHENIX | 2020-12-14 | 33786375 | - | - | - | - | N/A | - | - | 3C-like proteinase | 59.7 | - | No | - | 53.5 | 71.3 | 69.1 | - | ||||||
| 7l12 | 1.80 Å | 2021-03-03 | X-ray | XF4 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.11% | 24.91% | N/A | (5S)-5-{3-[3-(benzyloxy)-5-chlorophenyl]-2-oxo[2H-[1,3'-bipyridine]]-5-yl}pyrimidine-2,4(3H,5H)-dione | (5S)-5-{3-[3-(benzyloxy)-5-chlorophenyl]-2-oxo[2H-[1,3'-bipyridine]]-5-yl}pyrimidine-2,4(3H,5H)-dione | XDS | 95.20% | 8.27 | APS (24-ID-E) | PHENIX | 2020-12-14 | 33786375 | - | - | - | - | N/A | - | - | 3C-like proteinase | 44.8 | - | No | - | 36.0 | 56.0 | 73.2 | - | ||||||
| 7l13 | 2.17 Å | 2021-03-03 | X-ray | XF7 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.67% | 24.76% | N/A | (5S)-5-(3-{3-chloro-5-[(2-chlorophenyl)methoxy]phenyl}-2-oxo[2H-[1,3'-bipyridine]]-5-yl)pyrimidine-2,4(3H,5H)-dione | (5S)-5-(3-{3-chloro-5-[(2-chlorophenyl)methoxy]phenyl}-2-oxo[2H-[1,3'-bipyridine]]-5-yl)pyrimidine-2,4(3H,5H)-dione | XDS | 99.60% | 16.59 | NSLS-II (17-ID-1) | PHENIX | 2020-12-14 | 33786375 | - | - | - | - | N/A | - | - | 3C-like proteinase | 23.3 | - | No | - | 37.6 | 37.2 | 47.1 | - | ||||||
| 7l14 | 1.80 Å | 2021-03-03 | X-ray | XFD | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.57% | 20.44% | N/A | 2-{3-[3-chloro-5-(cyclopropylmethoxy)phenyl]-2-oxo[2H-[1,3'-bipyridine]]-5-yl}benzonitrile | 2-{3-[3-chloro-5-(cyclopropylmethoxy)phenyl]-2-oxo[2H-[1,3'-bipyridine]]-5-yl}benzonitrile | XDS | 95.00% | 14.94 | NSLS-II (17-ID-1) | PHENIX | 2020-12-14 | 33786375 | - | - | - | - | N/A | - | - | 3C-like proteinase | 71.0 | - | No | - | 78.6 | 52.3 | 86.9 | - | ||||||
| 7lop | 2.25 Å | 2021-03-03 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 21.97% | 26.80% | 14.60% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 98.90% | 4.3 | APS (23-ID-B) | PHENIX | 2021-02-10 | 34016740 | - | - | - | - | N/A | - | - | CV05-163 Fab light chain; CV05-163 Fab heavy chain; Spike protein S1; CR3022 Fab light chain; CR3022 Fab heavy chain | 18.4 | - | No | - | 20.8 | 26.7 | 63.5 | - | ||||||
| 7ltj | 1.80 Å | 2021-03-03 | X-ray | YD1 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.16% | 19.24% | 4.20% | 6-[4-(3,4-dichlorophenyl)piperazin-1-yl]carbonyl-1~{H}-pyrimidine-2,4-dione | 6-[4-(3,4-dichlorophenyl)piperazin-1-yl]carbonyl-1~{H}-pyrimidine-2,4-dione | CrysalisPro | 96.80% | 26.88 | ROTATING ANODE () | PHENIX | 2021-02-19 | 34793155 | - | - | - | - | N/A | - | - | 3C-like proteinase | 61.8 | - | No | - | 86.5 | 37.5 | 74.2 | - | ||||||
| 7nbr | 2.40 Å | 2021-03-03 | X-ray | U5G | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.01% | 26.94% | 9.30% | boceprevir (bound form) | boceprevir (bound form) | 99.90% | 15.6 | PETRA III, DESY (P11) | REFMAC | 2021-01-27 | - | - | - | - | - | N/A | - | - | Main Protease | 46.1 | - | No | - | - | 19.9 | 95.7 | 52.0 | - | ||||||
| 7nbs | 1.70 Å | 2021-03-03 | X-ray | SV6 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.61% | 25.60% | 7.30% | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide | 100.00% | 15.6 | PETRA III, DESY (P11) | REFMAC | 2021-01-27 | - | - | - | - | - | N/A | - | - | Main Protease | 29.6 | - | No | - | - | 29.8 | 48.7 | 56.5 | - | ||||||
| 7nby | 1.93 Å | 2021-03-03 | X-ray | NA, NO3, U88 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.44% | 19.82% | 10.70% | SODIUM ION; NITRATE ION; 5-nitro-1,3-thiazole | SODIUM ION; NITRATE ION; 5-nitro-1,3-thiazole | XDS | 99.80% | 11.0 | ELETTRA (11.2C) | PHENIX; PHENIX | 2021-01-28 | - | - | - | - | - | N/A | - | - | Main Protease | 73.8 | - | No | - | - | 82.9 | 49.8 | 92.3 | - | |||||
| 7nd3 | 3.70 Å | 2021-03-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-01-30 | 33756110 | - | - | - | - | N/A | - | - | COVOX-40 light chain; COVOX-40 heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7nd4 | 3.60 Å | 2021-03-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-01-30 | 33756110 | - | - | - | - | N/A | - | - | COVOX-88 Fab light chain; COVOX-88 Fab heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7nd5 | 3.40 Å | 2021-03-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-01-30 | 33756110 | - | - | - | - | N/A | - | - | COVOX-150 Fab light chain; COVOX-150 Fab heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7nd6 | 7.30 Å | 2021-03-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-01-30 | 33756110 | - | - | - | - | N/A | - | - | COVOX-158 Fab light chain; COVOX-158 Fab heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7nd7 | 3.60 Å | 2021-03-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-01-30 | 33756110 | - | - | - | - | N/A | - | - | COVOX-316 Fab light chain; COVOX-316 Fab heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7nd8 | 3.50 Å | 2021-03-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-01-30 | 33756110 | - | - | - | - | N/A | - | - | COVOX-384 Fab light chain; COVOX-384 Fab heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7nd9 | 2.80 Å | 2021-03-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-01-30 | 33756110 | - | - | - | - | N/A | - | - | COVOX-253H55L Fab light chain; COVOX-253H55L Fab heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7nda | 3.30 Å | 2021-03-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-01-30 | 33756110 | - | - | - | - | N/A | - | - | COVOX-253H55L Fab light chain; COVOX-253H55L Fab heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7ndb | 4.60 Å | 2021-03-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-01-30 | 33756110 | - | - | - | - | N/A | - | - | COVOX-253H165L Fab light chain; COVOX-253H165L Fab heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7ndc | 4.10 Å | 2021-03-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-01-30 | 33756110 | - | - | - | - | N/A | - | - | COVOX-159 Fab light chain; COVOX-159 heavy Fab chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7ndd | 4.20 Å | 2021-03-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-01-30 | 33756110 | - | - | - | - | N/A | - | - | COVOX-159 Fab light chain; COVOX-159 Fab heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7neg | 2.19 Å | 2021-03-03 | X-ray | - | SARS-CoV-2 | Spike | Protein-protein complex | 19.45% | 22.28% | 17.70% | - | xia2 | 99.90% | 8.4 | Diamond (I03) | PHENIX | 2021-02-04 | 33743891 | - | - | - | - | N/A | - | - | Surface glycoprotein; Antibody COVOX-269 Fab light chain; Antibody COVOX-269 Fab heavy chain | 52.3 | - | No | - | 62.5 | 41.5 | 75.5 | - | |||||||
| 7neh | 1.77 Å | 2021-03-03 | X-ray | NO3 | SARS-CoV-2 | Spike | Protein-protein complex | 18.82% | 19.82% | 9.10% | NITRATE ION | NITRATE ION | xia2 | 100.00% | 14.0 | Diamond (I03) | PHENIX | 2021-02-04 | 33743891 | - | - | - | - | N/A | - | - | Spike glycoprotein; COVOX-269 fab light chain; COVOX-269 Fab heavy chain | 57.4 | - | No | - | 82.9 | 37.4 | 69.1 | - | ||||||
| 7nev | 1.70 Å | 2021-03-03 | X-ray | IMD | SARS-CoV-2 | NSP5 (3CLpro) | Protein-protein complex | 18.32% | 23.36% | 6.70% | IMIDAZOLE; LEUPEPTIN | IMIDAZOLE | DIALS; XDS | 97.70% | 10.2 | PETRA III, DESY (P11) | PHENIX | 2021-02-05 | 33811162 | - | - | - | - | N/A | - | - | LEUPEPTIN; Main Protease | 69.5 | - | No | - | 51.5 | 73.6 | 89.4 | - | ||||||
| 5rob | 1.87 Å | 2021-03-10 | X-ray | PO4 | SARS-CoV-2 | NSP13 | PanDDA fragment screening | 21.87% | 25.38% | N/A | PHOSPHATE ION | PHOSPHATE ION | XDS | 96.00% | 8.2 | Diamond (I04-1) | REFMAC | 2020-09-22 | - | ZN | - | - | - | N/A | - | - | Helicase | 24.1 | - | No | - | - | 31.9 | 38.7 | 52.9 | - | |||||
| 7czp | 3.00 Å | 2021-03-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-09-09 | 33731853 | - | - | - | - | N/A | - | - | IG c181_light_IGLV2-14_IGLJ3,IGL@ protein; Immunoglobulin heavy variable 4-59,Chain H of P2B-1A1,Anti-RhD monoclonal T125 gamma1 heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7czq | 2.80 Å | 2021-03-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-09-09 | 33731853 | - | - | - | - | N/A | - | - | Immunoglobulin kappa variable 1-33,Uncharacterized protein; IG c642_heavy_IGHV3-53_IGHD1-26_IGHJ6,Chain H of P2B-1A10,Immunoglobulin gamma-1 heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7czr | 3.50 Å | 2021-03-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-09-09 | 33731853 | - | - | - | - | N/A | - | - | IGL c4203_light_IGKV1-9_IGKJ4,IGL c4203_light_IGKV1-9_IGKJ4,Uncharacterized protein; IG c542_heavy_IGHV3-53_IGHD3-10_IGHJ6,IGH@ protein; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7czs | 3.60 Å | 2021-03-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-09-09 | 33731853 | - | - | - | - | N/A | - | - | IGL c4203_light_IGKV1-9_IGKJ4,Uncharacterized protein; IG c542_heavy_IGHV3-53_IGHD3-10_IGHJ6,IGH@ protein; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7czt | 2.70 Å | 2021-03-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-09-09 | 33731853 | - | - | - | - | N/A | - | - | IG c689_light_IGLV5-37_IGLJ3,IGL@ protein; Immunoglobulin heavy variable 3-33,chainH of P5A-2G9,Immunoglobulin gamma-1 heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7czu | 3.40 Å | 2021-03-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-09-09 | 33731853 | - | - | - | - | N/A | - | - | Immunoglobulin kappa variable 1-33,Immunoglobulin kappa variable 1-33,Uncharacterized protein; Immunoglobulin heavy variable 3-30-3,Immunoglobulin heavy variable 3-30-3,Chain H of P5A-1B6_2B,Immunoglobulin gamma-1 heavy chain,Immunoglobulin gamma-1 heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7czv | 3.30 Å | 2021-03-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-09-09 | 33731853 | - | - | - | - | N/A | - | - | Immunoglobulin kappa variable 1-33,Uncharacterized protein; Immunoglobulin heavy variable 3-30-3,chain H of P5A-1B6_3B,Immunoglobulin gamma-1 heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7czw | 2.80 Å | 2021-03-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-09-09 | 33731853 | - | - | - | - | N/A | - | - | IGL c2312_light_IGLV2-14_IGLJ2,IGL@ protein; Immunoglobulin heavy variable 4-61,chain H of P5A-2G7,Epididymis luminal protein 214; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7czx | 2.80 Å | 2021-03-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-09-09 | 33731853 | - | - | - | - | N/A | - | - | IG c168_light_IGKV4-1_IGKJ4,Uncharacterized protein; Immunoglobulin heavy variable 4-59,chain H of P5A-1B9,IGH@ protein; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7czy | 3.30 Å | 2021-03-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-09-09 | 33731853 | - | - | - | - | N/A | - | - | IG c168_light_IGKV4-1_IGKJ4,Immunoglobulin kappa constant; Immunoglobulin heavy variable 1-8,Immunoglobulin heavy variable 1-8,chain H of P5A-2F11_2B,Epididymis luminal protein 214,Epididymis luminal protein 214; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7czz | 3.20 Å | 2021-03-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-09-09 | 33731853 | - | - | - | - | N/A | - | - | IG c168_light_IGKV4-1_IGKJ4,Immunoglobulin kappa constant; Immunoglobulin heavy variable 1-8,Immunoglobulin heavy variable 1-8,chain H of P5A-2F11_3B,Epididymis luminal protein 214,Epididymis luminal protein 214; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7d00 | 3.00 Å | 2021-03-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-09-09 | 33731853 | - | - | - | - | N/A | - | - | IGL c4203_light_IGKV1-9_IGKJ4,Uncharacterized protein; IG c642_heavy_IGHV3-53_IGHD1-26_IGHJ6,chain H of FabP5A-1B8,IGH@ protein; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7d03 | 3.20 Å | 2021-03-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-09-09 | 33731853 | - | - | - | - | N/A | - | - | IGL c2312_light_IGLV2-14_IGLJ2,IGL@ protein; Immunoglobulin heavy variable 4-61,chain H of FabP5A-2G7,Epididymis luminal protein 214; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7d0b | 3.90 Å | 2021-03-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-09-09 | 33731853 | - | - | - | - | N/A | - | - | Light chain of P5A-3C12; Heavy chain of P5A-3C12; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7d0c | 3.40 Å | 2021-03-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-09-09 | 33731853 | - | - | - | - | N/A | - | - | Light chain of P5A-3A1; Heavy chain of P5A-3A1; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7d0d | 3.80 Å | 2021-03-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-09-09 | 33731853 | - | - | - | - | N/A | - | - | Light chain of P5A-3C12; Heavy chain of P5A-3C12; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7jsu | 1.83 Å | 2021-03-10 | X-ray | PO4, UED | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.93% | 20.28% | N/A | PHOSPHATE ION; N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | PHOSPHATE ION; N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | XDS | 99.00% | 12.7 | APS (24-ID-C) | PHENIX | 2020-08-16 | 33795671 | - | - | - | - | N/A | - | - | 3C-like proteinase | 64.5 | - | No | - | 79.8 | 52.0 | 72.0 | - | ||||||
| 7jt0 | 1.73 Å | 2021-03-10 | X-ray | PO4, LW1 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.66% | 19.17% | 4.00% | PHOSPHATE ION; thiophene-2-carbaldehyde | PHOSPHATE ION; thiophene-2-carbaldehyde | XDS | 99.20% | 23.5 | APS (24-ID-E) | PHENIX | 2020-08-16 | 33795671 | - | - | - | - | N/A | - | - | 3C-like proteinase | 70.2 | - | No | - | 86.9 | 42.1 | 87.2 | - | ||||||
| 7jw8 | 1.84 Å | 2021-03-10 | X-ray | BTB, TG3 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.31% | 22.72% | 11.00% | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL; ethyl (4R)-4-[[(2S)-4-methyl-2-[[(2S,3R)-3-[(2-methylpropan-2-yl)oxy]-2-(phenylmethoxycarbonylamino)butanoyl]amino]pentanoyl]amino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL; ethyl (4R)-4-[[(2S)-4-methyl-2-[[(2S,3R)-3-[(2-methylpropan-2-yl)oxy]-2-(phenylmethoxycarbonylamino)butanoyl]amino]pentanoyl]amino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | XDS | 97.60% | 5.7 | APS (24-ID-C) | PHENIX | 2020-08-25 | 33795671 | - | - | - | - | N/A | - | - | 3C-like proteinase | 69.4 | - | No | - | 57.9 | 86.1 | 70.3 | - | ||||||
| 7lab | 2.97 Å | 2021-03-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-01-06 | 34242577 | - | - | - | - | N/A | - | - | Spike glycoprotein; DH1052 heavy chain - Receptor Binding Domain; DH1052 light chain - Receptor Binding Domain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 6yxj | 3.50 Å | 2021-03-17 | X-ray | - | SARS-CoV | NSP3 | Protein-protein complex | 31.27% | 33.55% | 9.50% | - | XDS | 99.30% | 10.8 | BESSY, BESSY (14.2, 14.2) | BUSTER | 2020-05-02 | 33876849 | - | - | - | - | N/A | - | - | Polyadenylate-binding protein-interacting protein 1; Non-structural protein 3 | 24.0 | - | No | - | 4.2 | 89.9 | 29.3 | - | |||||||
| 7e35 | 2.40 Å | 2021-03-17 | X-ray | GYX | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 28.14% | 31.71% | 17.20% | N-[(3-acetamidophenyl)methyl]-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide | N-[(3-acetamidophenyl)methyl]-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide | XDS | 92.20% | 19.1 | SSRF (BL17U1) | PHENIX; PHENIX | 2021-02-08 | 33979649 | ZN | - | - | - | N/A | - | - | Non-structural protein 3 - papain-like protease (PLPro) | 1.1 | - | No | - | 5.2 | 13.5 | 27.5 | - | ||||||
| 7eam | 1.40 Å | 2021-03-17 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 17.20% | 18.17% | 6.10% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | Aimless | 94.70% | 8.8 | SSRF (BL17U1) | PHENIX | 2021-03-07 | - | - | - | - | - | N/A | - | - | the light chain of Fab fragment of antibody 7D6; the heavy chain of Fab fragment of antibody 7D6; Spike protein S1 - UNP residues 319-541 | 71.0 | - | No | - | - | 91.9 | 59.2 | 66.8 | - | |||||
| 7jt7 | 1.94 Å | 2021-03-17 | X-ray | TG3 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.42% | 22.54% | N/A | ethyl (4R)-4-[[(2S)-4-methyl-2-[[(2S,3R)-3-[(2-methylpropan-2-yl)oxy]-2-(phenylmethoxycarbonylamino)butanoyl]amino]pentanoyl]amino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | ethyl (4R)-4-[[(2S)-4-methyl-2-[[(2S,3R)-3-[(2-methylpropan-2-yl)oxy]-2-(phenylmethoxycarbonylamino)butanoyl]amino]pentanoyl]amino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | XDS | 99.80% | 10.2 | APS (24-ID-C) | PHENIX | 2020-08-17 | 33795671 | - | - | - | - | N/A | - | - | 3C-like proteinase | 28.0 | - | No | - | 59.8 | 14.1 | 57.9 | - | ||||||
| 7laa | 3.42 Å | 2021-03-17 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-01-06 | 34242577 | - | - | - | - | N/A | - | - | DH1041 light chain - Receptor Binding Domain; DH1041 heavy chain - Receptor Binding Domain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7ls9 | 3.42 Å | 2021-03-17 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-02-17 | 34111408 | - | - | - | - | N/A | - | - | 1-57 Fab light chain; 1-57 Fab heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lss | 3.72 Å | 2021-03-17 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-02-18 | 34111408 | - | - | - | - | N/A | - | - | Fab 2-7 variable heavy chain; Fab 2-7 variable light chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lyh | 1.90 Å | 2021-03-17 | X-ray | YHJ | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.99% | 22.19% | 11.30% | benzyl (1S,3aR,6aS)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamoyl)hexahydrocyclopenta[c]pyrrole-2(1H)-carboxylate | benzyl (1S,3aR,6aS)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamoyl)hexahydrocyclopenta[c]pyrrole-2(1H)-carboxylate | Aimless | 96.20% | 8.7 | APS (19-ID) | REFMAC | 2021-03-07 | 34414360 | - | - | - | - | N/A | - | - | 3C-like proteinase | 70.5 | - | No | - | 63.4 | 83.5 | 69.9 | - | ||||||
| 7lyi | 1.90 Å | 2021-03-17 | X-ray | YHI, NA | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.03% | 21.73% | 6.90% | benzyl (1R,2S,5S)-2-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamoyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-3-carboxylate; SODIUM ION | benzyl (1R,2S,5S)-2-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamoyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-3-carboxylate; SODIUM ION | Aimless | 93.00% | 6.5 | APS (19-ID) | REFMAC | 2021-03-07 | 34414360 | - | - | - | - | N/A | - | - | 3C-like proteinase | 69.6 | - | No | - | 67.7 | 68.7 | 78.3 | - | ||||||
| 7nio | 2.20 Å | 2021-03-17 | X-ray | - | SARS-CoV-2 | NSP13 | No functional ligands | 22.96% | 28.65% | N/A | - | XDS | 90.30% | 4.4 | Diamond (I04-1) | PHENIX | 2021-02-12 | 34381037 | ZN | - | - | - | N/A | - | - | SARS-CoV-2 helicase NSP13 | 14.8 | - | No | - | 11.3 | 50.0 | 40.8 | - | |||||||
| 6w81 | 1.55 Å | 2021-03-24 | X-ray | MLA, X77 | PEDV | NSP5 (3CLpro) | Functional ligand | 17.58% | 21.36% | N/A | MALONIC ACID; N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide | MALONIC ACID; N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide | HKL-2000 | 97.80% | 28.9 | APS (21-ID-G) | PHENIX | 2020-03-20 | - | - | - | - | - | N/A | - | - | Peptidase C30 | 28.7 | - | No | - | - | 71.0 | 28.2 | 34.0 | - | |||||
| 7cr5 | 2.08 Å | 2021-03-24 | X-ray | - | SARS-CoV-2 | Nucleocapsid | Pathogen-host interaction | 19.08% | 22.20% | 22.00% | - | HKL-3000 | 99.90% | 13.18 | SSRF (BL18U1) | PHENIX; PHENIX | 2020-08-12 | 33976229 | ZN | - | - | - | N/A | - | - | monoclonal antibody chain L; monoclonal antibody chain H; Nucleoprotein | 53.5 | - | No | - | 63.1 | 43.6 | 75.1 | - | |||||||
| 7dcd | 2.57 Å | 2021-03-24 | X-ray | - | SARS-CoV-2 | NSP7/NSP8 | Protein-protein complex | 25.39% | 35.33% | 20.00% | - | 93.15% | 8.5 | SSRF (BL18U1) | PHENIX | 2020-10-24 | 33594727 | - | - | - | - | N/A | - | - | Non-structural protein 8; Non-structural protein 7 | 1.3 | - | No | - | 3.8 | 31.9 | 12.5 | - | ||||||||
| 7e7b | 2.60 Å | 2021-03-24 | Cryo-EM | VCG, ELA, NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-hydroxyethyl 2-deoxy-3,5-bis-O-(2-hydroxyethyl)-6-O-(2-{[(9E)-octadec-9-enoyl]oxy}ethyl)-alpha-L-xylo-hexofuranoside; 9-OCTADECENOIC ACID; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-hydroxyethyl 2-deoxy-3,5-bis-O-(2-hydroxyethyl)-6-O-(2-{[(9E)-octadec-9-enoyl]oxy}ethyl)-alpha-L-xylo-hexofuranoside; 9-OCTADECENOIC ACID; 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-02-25 | 33692215 | - | - | - | - | N/A | - | - | Spike glycoprotein,Collagen alpha-1(I) chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7e7d | 3.20 Å | 2021-03-24 | Cryo-EM | ELA, NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 9-OCTADECENOIC ACID; 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 9-OCTADECENOIC ACID; 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-02-25 | 33692215 | - | - | - | - | N/A | - | - | Spike glycoprotein,Collagen alpha-1(I) chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7krr | 3.50 Å | 2021-03-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | 43.51% | 43.66% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-11-20 | 33727252 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7krs | 3.20 Å | 2021-03-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | 47.23% | 47.92% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-11-20 | 33727252 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7l2d | 3.55 Å | 2021-03-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-12-16 | 33789084 | - | - | - | - | N/A | - | - | 1-87 light chain; 1-87 heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7l2e | 2.97 Å | 2021-03-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-12-16 | 33789084 | - | - | - | - | N/A | - | - | 4-18 light chain; 4-18 heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7l2f | 3.90 Å | 2021-03-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-12-16 | 33789084 | - | - | - | - | N/A | - | - | 5-24 light chain; 5-24 heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lqv | 3.25 Å | 2021-03-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-02-15 | - | - | - | - | - | N/A | - | - | 4-8 Light chain; 4-8 Heavy Chain; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7lqw | 4.47 Å | 2021-03-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-02-15 | 33789084 | - | - | - | - | N/A | - | - | Spike glycoprotein; 2-17 Light Chain; 2-17 Heavy Chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7lzt | 1.55 Å | 2021-03-24 | X-ray | YN1, YMY | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.79% | 19.66% | 9.30% | (1R,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1s,4S)-4-propylcyclohexyl)methoxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | (1S,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1s,4S)-4-propylcyclohexyl)methoxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1R,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1s,4S)-4-propylcyclohexyl)methoxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | XDS | 100.00% | 10.5 | APS (17-ID) | PHENIX | 2021-03-10 | 34865476 | - | - | - | - | N/A | - | - | 3C-like proteinase | 84.4 | - | No | - | 84.0 | 88.4 | 89.8 | - | ||||||
| 7lzu | 1.60 Å | 2021-03-24 | X-ray | PG4, YKP, YKM | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.78% | 20.11% | 5.40% | (1S,2S)-2-((S)-2-((((S)-1-(4,4-difluorocyclohexyl)ethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1R,2S)-2-((S)-2-((((S)-1-(4,4-difluorocyclohexyl)ethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | TETRAETHYLENE GLYCOL; (1S,2S)-2-((S)-2-((((S)-1-(4,4-difluorocyclohexyl)ethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1R,2S)-2-((S)-2-((((S)-1-(4,4-difluorocyclohexyl)ethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | XDS | 97.80% | 12.5 | APS (17-ID) | PHENIX | 2021-03-10 | 34865476 | - | - | - | - | N/A | - | - | 3C-like proteinase | 73.5 | - | No | - | 80.9 | 55.0 | 88.0 | - | ||||||
| 7lzv | 1.60 Å | 2021-03-24 | X-ray | PG4, YLS, YLM | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.46% | 21.19% | 4.10% | (1S,2S)-2-((S)-2-((((4-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1R,2S)-2-((S)-2-((((4-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | TETRAETHYLENE GLYCOL; (1S,2S)-2-((S)-2-((((4-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1R,2S)-2-((S)-2-((((4-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | XDS | 98.90% | 14.7 | APS (17-ID) | PHENIX | 2021-03-10 | 34865476 | - | - | - | - | N/A | - | - | 3C-like proteinase | 66.2 | - | No | - | 72.6 | 47.8 | 87.0 | - | ||||||
| 7lzw | 2.20 Å | 2021-03-24 | X-ray | YLS, YLM | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.35% | 25.76% | 10.10% | (1R,2S)-2-((S)-2-((((4-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | (1S,2S)-2-((S)-2-((((4-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1R,2S)-2-((S)-2-((((4-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | XDS | 99.70% | 7.9 | APS (17-ID) | PHENIX | 2021-03-10 | 34865476 | - | - | - | - | N/A | - | - | 3C-like proteinase | 68.1 | - | No | - | 28.6 | 95.0 | 87.8 | - | ||||||
| 7lzx | 1.65 Å | 2021-03-24 | X-ray | PG4, YMD, YMG | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.48% | 21.39% | 5.30% | (1R,2S)-2-((S)-2-((((4,4-dimethylcyclohexyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1S,2S)-2-((S)-2-((((4,4-dimethylcyclohexyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | TETRAETHYLENE GLYCOL; (1R,2S)-2-((S)-2-((((4,4-dimethylcyclohexyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1S,2S)-2-((S)-2-((((4,4-dimethylcyclohexyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | XDS | 99.20% | 10.7 | APS (17-ID) | PHENIX | 2021-03-10 | 34865476 | - | - | - | - | N/A | - | - | 3C-like proteinase | 69.0 | - | No | - | 70.8 | 53.5 | 89.2 | - | ||||||
| 7lzy | 1.85 Å | 2021-03-24 | X-ray | YMM, YMJ | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.57% | 22.29% | 8.50% | (1R,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1s,4S)-4-propylcyclohexyl)oxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | (1S,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1s,4S)-4-propylcyclohexyl)oxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1R,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1s,4S)-4-propylcyclohexyl)oxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | XDS | 100.00% | 11.4 | APS (17-ID) | PHENIX | 2021-03-10 | 34865476 | - | - | - | - | N/A | - | - | 3C-like proteinase | 80.7 | - | No | - | 62.4 | 93.3 | 89.6 | - | ||||||
| 7lzz | 2.00 Å | 2021-03-24 | X-ray | YMV, YMS | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.23% | 22.90% | 9.80% | (1R,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1r,4S)-4-phenylcyclohexyl)oxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | (1S,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1r,4S)-4-phenylcyclohexyl)oxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1R,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1r,4S)-4-phenylcyclohexyl)oxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | XDS | 99.90% | 7.5 | APS (17-ID) | PHENIX | 2021-03-10 | 34865476 | - | - | - | - | N/A | - | - | 3C-like proteinase | 79.1 | - | No | - | 55.9 | 95.7 | 88.3 | - | ||||||
| 7m00 | 2.00 Å | 2021-03-24 | X-ray | YKD, YKA | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 21.13% | 26.54% | 9.40% | (1R,2S)-2-((S)-2-((((2-(4,4-difluorocyclohexyl)propan-2-yl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl)propane-1-sulfonic acid | (1S,2S)-2-((S)-2-((((2-(4,4-difluorocyclohexyl)propan-2-yl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl)propane-1-sulfonic acid; (1R,2S)-2-((S)-2-((((2-(4,4-difluorocyclohexyl)propan-2-yl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl)propane-1-sulfonic acid | XDS | 100.00% | 8.9 | APS (17-ID) | PHENIX | 2021-03-10 | 34865476 | - | - | - | - | N/A | - | - | 3C-like proteinase | 60.8 | - | No | - | 22.7 | 95.7 | 77.8 | - | ||||||
| 7m01 | 1.65 Å | 2021-03-24 | X-ray | YKS, YKV | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.80% | 22.39% | 13.90% | (1S,2S)-2-((S)-2-((((S)-1-(4,4-difluorocyclohexyl)-2-phenylethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | (1R,2S)-2-((S)-2-((((S)-1-(4,4-difluorocyclohexyl)-2-phenylethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1S,2S)-2-((S)-2-((((S)-1-(4,4-difluorocyclohexyl)-2-phenylethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | XDS | 100.00% | 8.7 | NSLS-II (17-ID-1) | PHENIX | 2021-03-10 | 34865476 | - | - | - | - | N/A | - | - | 3C-like proteinase | 67.5 | - | No | - | 61.4 | 63.5 | 85.3 | - | ||||||
| 7m02 | 1.80 Å | 2021-03-24 | X-ray | PG4, YKY, YL7 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.10% | 24.07% | 8.00% | (1R,2S)-2-((S)-2-((((2-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl)propane-1-sulfonic acid; (1S,2S)-2-((S)-2-((((2-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl)propane-1-sulfonic acid | TETRAETHYLENE GLYCOL; (1R,2S)-2-((S)-2-((((2-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl)propane-1-sulfonic acid; (1S,2S)-2-((S)-2-((((2-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl)propane-1-sulfonic acid | XDS | 99.70% | 10.3 | APS (17-ID) | PHENIX | 2021-03-10 | 34865476 | - | - | - | - | N/A | - | - | 3C-like proteinase | 71.0 | - | No | - | 44.3 | 83.2 | 90.5 | - | ||||||
| 7m03 | 2.00 Å | 2021-03-24 | X-ray | YLJ, YLD | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.30% | 24.88% | 6.40% | (1R,2S)-2-((S)-2-((((3-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | (1S,2S)-2-((S)-2-((((3-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid; (1R,2S)-2-((S)-2-((((3-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid | XDS | 97.40% | 10.3 | APS (17-ID) | PHENIX | 2021-03-10 | 34865476 | - | - | - | - | N/A | - | - | 3C-like proteinase | 63.9 | - | No | - | 36.5 | 88.4 | 77.6 | - | ||||||
| 7m04 | 1.75 Å | 2021-03-24 | X-ray | PG4, YM1, YLV | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.99% | 22.28% | 5.70% | (1S,2S)-1-hydroxy-2-((S)-4-methyl-2-((((perfluorophenyl)methoxy)carbonyl)amino)pentanamido)-3-((R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl)propane-1-sulfonic acid; (1R,2S)-1-hydroxy-2-((S)-4-methyl-2-((((perfluorophenyl)methoxy)carbonyl)amino)pentanamido)-3-((R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl)propane-1-sulfonic acid | TETRAETHYLENE GLYCOL; (1S,2S)-1-hydroxy-2-((S)-4-methyl-2-((((perfluorophenyl)methoxy)carbonyl)amino)pentanamido)-3-((R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl)propane-1-sulfonic acid; (1R,2S)-1-hydroxy-2-((S)-4-methyl-2-((((perfluorophenyl)methoxy)carbonyl)amino)pentanamido)-3-((R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl)propane-1-sulfonic acid | XDS | 99.70% | 12.2 | APS (17-ID) | PHENIX | 2021-03-10 | 34865476 | - | - | - | - | N/A | - | - | 3C-like proteinase | 74.0 | - | No | - | 62.5 | 83.2 | 79.6 | - | ||||||
| 7m1y | 2.02 Å | 2021-03-24 | X-ray | NA, FMT, IOD, 9JT | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 18.41% | 21.29% | 15.00% | SODIUM ION; FORMIC ACID; IODIDE ION; ~{N}-phenyl-2-selanyl-benzamide | SODIUM ION; FORMIC ACID; IODIDE ION; ~{N}-phenyl-2-selanyl-benzamide | HKL-3000 | 100.00% | 5.0 | APS (19-ID) | REFMAC | 2021-03-15 | - | NA; ZN | - | - | - | N/A | - | - | Papain-like protease | 74.6 | - | No | - | - | 71.7 | 81.6 | 73.6 | - | |||||
| 7nn0 | 3.04 Å | 2021-03-24 | X-ray | ANP | SARS-CoV-2 | NSP13 | No functional ligands | 24.38% | 28.38% | 25.80% | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | DIALS | 97.70% | 6.1 | Diamond (I04-1) | PHENIX | 2021-02-23 | 34381037 | MG; ZN | - | - | - | N/A | - | - | SARS-CoV-2 helicase NSP13 | 26.7 | - | No | - | 12.5 | 76.0 | 40.5 | - | ||||||
| 7deo | 2.50 Å | 2021-03-31 | X-ray | NAG, CA | SARS-CoV-2 | Spike | Pathogen-host interaction | 18.72% | 23.21% | 12.80% | 2-acetamido-2-deoxy-beta-D-glucopyranose; CALCIUM ION | 2-acetamido-2-deoxy-beta-D-glucopyranose; CALCIUM ION | MOSFLM | 99.40% | 7.2 | SwissFEL ARAMIS (ESA) | PHENIX | 2020-11-04 | 33961621 | CA | - | - | - | N/A | - | - | Spike protein S1 - RBD; antibody scFv | 65.6 | - | No | - | 52.9 | 82.9 | 70.4 | - | ||||||
| 7det | 2.20 Å | 2021-03-31 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 21.20% | 25.99% | 11.20% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.10% | 18.6 | SSRF (BL17U) | PHENIX | 2020-11-05 | 33961621 | - | - | - | - | N/A | - | - | antibody scFv; Spike protein S1 - RBD | 30.7 | - | No | - | 26.8 | 53.3 | 57.4 | - | ||||||
| 7deu | 2.10 Å | 2021-03-31 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 20.86% | 22.94% | 9.00% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | MOSFLM | 100.00% | 23.8 | SwissFEL ARAMIS (ESA) | PHENIX | 2020-11-05 | 33961621 | - | - | - | - | N/A | - | - | antibody scFv; Spike protein S1 - RBD | 40.2 | - | No | - | 55.6 | 32.6 | 67.9 | - | ||||||
| 7dwx | 8.30 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | LEUCINE; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-01-18 | 33737693 | ZN | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2; Sodium-dependent neutral amino acid transporter B(0)AT1 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7dwy | 2.70 Å | 2021-03-31 | Cryo-EM | EIC, NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | LINOLEIC ACID; 2-acetamido-2-deoxy-beta-D-glucopyranose | LINOLEIC ACID; 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-01-18 | 33737693 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7dwz | 3.30 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-01-18 | 33737693 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7dx0 | 3.20 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-01-18 | 33737693 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7dx1 | 3.10 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-01-18 | 33737693 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7dx2 | 3.30 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-01-18 | 33737693 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7dx3 | 3.50 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-01-18 | 33737693 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7dx5 | 3.30 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-01-18 | 33737693 | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7dx6 | 3.00 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-01-18 | 33737693 | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7dx7 | 3.40 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-01-18 | 33737693 | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7dx8 | 2.90 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-01-18 | 33737693 | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7dx9 | 3.60 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-01-18 | 33737693 | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ean | 1.91 Å | 2021-03-31 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 19.11% | 22.39% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 99.80% | 13.7 | SSRF (BL17U1) | PHENIX | 2021-03-07 | 34580306 | - | - | - | - | N/A | - | - | Light chain of SARS-CoV-2 cross-neutralizing mAb 6D6; Heavy chain of SARS-CoV-2 cross-neutralizing mAb 6D6; Spike protein S1 | 31.5 | - | No | - | 61.4 | 16.1 | 61.5 | - | ||||||
| 7krq | 3.44 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | 42.23% | 42.48% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-11-20 | 33727252 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7ljr | 3.66 Å | 2021-03-31 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | 2021-01-30 | 33442694 | - | - | - | - | N/A | - | - | Fab DH1043 light chain; Fab DH1043 heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7lm8 | 1.94 Å | 2021-03-31 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 16.87% | 20.07% | 8.60% | - | HKL-2000 | 100.00% | 6.0 | APS (23-ID-D) | PHENIX | 2021-02-05 | 33894127 | - | - | - | - | N/A | - | - | CV38-142 Fab light chain; CV38-142 Fab heavy chain; COVA1-16 Fab light chain; COVA1-16 Fab heavy chain; Spike protein S1 | 73.7 | - | No | - | 81.3 | 63.8 | 79.4 | - | |||||||
| 7lm9 | 1.53 Å | 2021-03-31 | X-ray | - | SARS-CoV | Spike | Pathogen-host interaction | 16.97% | 19.22% | 9.10% | - | HKL-2000 | 95.70% | 7.0 | APS (23-ID-B) | PHENIX | 2021-02-05 | 33894127 | - | - | - | - | N/A | - | - | CV38-142 Fab light chain; CV38-142 Fab heavy chain; Spike glycoprotein | 69.8 | - | No | - | 86.6 | 51.9 | 76.9 | - | |||||||
| 7lo4 | 2.46 Å | 2021-03-31 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 19.28% | 25.14% | 16.20% | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.70% | 14.1 | Australian Synchrotron (MX2) | PHENIX | 2021-02-09 | - | ZN | - | - | - | N/A | - | - | Spike protein S1; Processed angiotensin-converting enzyme 2 | 50.8 | - | No | - | - | 33.9 | 61.5 | 81.2 | - | |||||
| 7lwi | 3.07 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-03-01 | 34168071 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lwj | 3.24 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-03-01 | 34168071 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lwk | 2.92 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-03-01 | 34168071 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lwl | 2.84 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-03-01 | 34168071 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lwm | 2.83 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-03-01 | 34168071 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lwn | 2.94 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-03-01 | 34168071 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lwo | 2.85 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-03-01 | 34168071 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lwp | 3.01 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-03-01 | 34168071 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lwq | 3.44 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-03-01 | 34168071 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7lws | 3.22 Å | 2021-03-31 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2021-03-01 | 33758838 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7lwt | 3.19 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-03-01 | 34168071 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lwu | 3.22 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-03-01 | 34168071 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lwv | 3.12 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-03-01 | 34168071 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lww | 3.00 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-03-01 | 34168071 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lyk | 3.65 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-03-07 | 34168071 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lyl | 3.72 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-03-07 | 34168071 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lym | 3.57 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-03-07 | 34168071 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lyn | 3.32 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-03-07 | 34168071 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lyo | 3.32 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-03-07 | 34168071 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lyp | 4.05 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-03-07 | 34168071 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lyq | 3.34 Å | 2021-03-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-03-07 | 34168071 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7m0j | 3.52 Å | 2021-03-31 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2021-03-11 | 33758838 | - | - | - | - | N/A | - | - | Spike glycoprotein - ectodomain (UNP residues 16-1208) | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7ca8 | 2.45 Å | 2021-04-07 | X-ray | FNO | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 22.95% | 27.89% | 12.80% | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione | XDS | 99.70% | 15.7 | SSRF (BL17U1) | PHENIX; PHENIX | 2020-06-08 | 33163253 | - | - | - | - | N/A | - | - | 3C-like proteinase | 26.3 | - | No | - | 14.7 | 71.9 | 41.5 | - | ||||||
| 7d4g | 3.90 Å | 2021-04-07 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | 2020-09-23 | 34676098 | - | - | - | - | N/A | - | - | Heavy chain of FC05 Fab; Light chain of FC05 Fab; Spike glycoprotein S1 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7e3j | 2.99 Å | 2021-04-07 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 22.72% | 24.79% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 98.50% | 8.8 | SSRF (BL19U1) | PHENIX | 2021-02-08 | 34234119 | ZN | - | - | - | N/A | - | - | Spike protein S1; ACE2 | 38.2 | - | No | - | 37.4 | 63.4 | 51.6 | - | |||||||
| 7mbg | 1.86 Å | 2021-04-07 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 19.49% | 22.21% | 17.60% | - | XDS | 99.60% | 7.2 | Diamond (I04) | PHENIX | 2021-03-31 | 34174328 | - | - | - | - | N/A | - | - | 3C-like proteinase | 46.7 | - | No | - | 63.0 | 57.7 | 48.0 | - | |||||||
| 7nf5 | 1.94 Å | 2021-04-07 | X-ray | NA, IPA, ALD | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 18.19% | 22.46% | 14.60% | SODIUM ION; ISOPROPYL ALCOHOL; N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | SODIUM ION; ISOPROPYL ALCOHOL; N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | XDS | 100.00% | 8.0 | ELETTRA (11.2C) | PHENIX; PHENIX | 2021-02-05 | 34769210 | - | - | - | - | N/A | - | - | Main Protease | 68.3 | - | No | - | 60.7 | 68.7 | 82.6 | - | ||||||
| 7ng3 | 1.80 Å | 2021-04-07 | X-ray | ALD | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 17.87% | 21.47% | 7.10% | N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | XDS | 96.60% | 10.4 | ELETTRA (11.2C) | PHENIX; PHENIX | 2021-02-08 | 34769210 | - | - | - | - | N/A | - | - | Main Protease | 66.8 | - | No | - | 70.1 | 64.3 | 74.2 | - | ||||||
| 7ng6 | 1.87 Å | 2021-04-07 | X-ray | ALD | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 16.19% | 20.77% | 7.10% | N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | XDS | 96.80% | 8.5 | ELETTRA (11.2C) | PHENIX; PHENIX | 2021-02-08 | 34769210 | - | - | - | - | N/A | - | - | Main Protease | 67.5 | - | No | - | 76.0 | 53.9 | 80.4 | - | ||||||
| 7nng | 2.38 Å | 2021-04-07 | X-ray | PO4, UJK | SARS-CoV-2 | NSP13 | Functional ligand | 24.48% | 29.48% | 16.30% | PHOSPHATE ION; 1-(2-Methylphenyl)-1H-1,2,3-triazole-4-carboxylic acid | PHOSPHATE ION; 1-(2-Methylphenyl)-1H-1,2,3-triazole-4-carboxylic acid | XDS | 96.60% | 4.1 | Diamond (I04-1) | PHENIX | 2021-02-24 | 34381037 | ZN | - | - | - | N/A | - | - | SARS-CoV-2 helicase NSP13 | 8.8 | - | No | - | 8.6 | 15.3 | 60.7 | - | ||||||
| 7nx6 | 2.25 Å | 2021-04-07 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 20.73% | 24.84% | 43.80% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 99.80% | 4.3 | Diamond (I03) | PHENIX | 2021-03-17 | 33852911 | - | - | - | - | N/A | - | - | COVOX-222 Fab light chain; COVOX-222 Fab Heavy chain; Spike protein S1; EY6A Fab light chain; EY6A Fab heavy chain | 28.2 | - | No | - | 36.7 | 16.9 | 78.5 | - | ||||||
| 7nx7 | 2.30 Å | 2021-04-07 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 20.75% | 23.90% | 24.50% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 100.00% | 7.7 | Diamond (I03) | PHENIX | 2021-03-17 | 33852911 | - | - | - | - | N/A | - | - | COVOX-222 Fab light chain; COVOX-222 Fab heavy chain; Spike protein S1; EY6A Fab light chain; EY6A Fab heavy chain | 27.1 | - | No | - | 45.7 | 12.3 | 71.7 | - | ||||||
| 7nx8 | 1.95 Å | 2021-04-07 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 22.32% | 24.58% | 12.00% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 100.00% | 12.0 | Diamond (I03) | PHENIX | 2021-03-17 | 33852911 | - | - | - | - | N/A | - | - | COVOX-222 Fab light chain; COVOX-222 Fab heavy chain; Spike protein S1; EY6A Fab light chain; EY6A Fab heavy chain | 25.3 | - | No | - | 39.5 | 11.6 | 74.9 | - | ||||||
| 7nx9 | 2.40 Å | 2021-04-07 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 19.95% | 22.85% | 25.70% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 100.00% | 7.7 | Diamond (I03) | PHENIX | 2021-03-17 | 33852911 | - | - | - | - | N/A | - | - | COVOX-222 Fab light chain; COVOX-222 Fab heavy chain; Spike protein S1; EY6A Fab light chain; EY6A Fab heavy chain | 40.9 | - | No | - | 56.6 | 21.7 | 79.5 | - | ||||||
| 7nxa | 2.50 Å | 2021-04-07 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 20.54% | 25.25% | 40.10% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 99.90% | 4.6 | Diamond (I03) | PHENIX | 2021-03-17 | 33852911 | - | - | - | - | N/A | - | - | COVOX-222 Fab light chain; COVOX-222 Fab heavy chain; Spike protein S1; EY6A Fab light chain; EY6A Fab heavy chain | 26.8 | - | No | - | 33.0 | 18.1 | 78.0 | - | ||||||
| 7nxb | 2.67 Å | 2021-04-07 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 21.26% | 24.60% | 36.50% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 100.00% | 7.0 | Diamond (I03) | PHENIX | 2021-03-17 | 33852911 | - | - | - | - | N/A | - | - | COVOX-222 Fab light chain; COVOX-222 Fab heavy chain; Spike protein S1; EY6A Fab light chain; EY6A Fab heavy chain | 27.0 | - | No | - | 39.0 | 14.2 | 76.3 | - | ||||||
| 7nxc | 3.14 Å | 2021-04-07 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 22.63% | 27.83% | 50.00% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 100.00% | 5.3 | Diamond (I03) | PHENIX | 2021-03-17 | 33852911 | - | - | - | - | N/A | - | - | Spike protein S1; Processed angiotensin-converting enzyme 2 | 21.1 | - | No | - | 14.9 | 34.7 | 67.4 | - | ||||||
| 7l56 | 3.60 Å | 2021-04-14 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-12-21 | 33794145 | - | - | - | - | N/A | - | - | Fab 2-43 variable domain light chain; Fab 2-43 variable domain heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7l57 | 5.87 Å | 2021-04-14 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-12-21 | 33794145 | - | - | - | - | N/A | - | - | Fab 2-15 variable domain light chain; Fab 2-15 variable domain heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7l58 | 5.07 Å | 2021-04-14 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-12-21 | 33794145 | - | - | - | - | N/A | - | - | Fab H4 variable domain light chain; Fab H4 variable domain heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lkd | 2.01 Å | 2021-04-14 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 18.24% | 22.57% | 8.00% | - | HKL-2000 | 99.80% | 21.5 | CHESS (A1) | PHENIX | 2021-02-02 | 35217718 | - | - | - | - | N/A | - | - | 3C-like proteinase | 62.7 | - | No | - | 59.6 | 71.3 | 69.1 | - | |||||||
| 7lke | 2.69 Å | 2021-04-14 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 22.51% | 29.37% | 10.40% | - | HKL-2000 | 92.80% | 11.6 | CHESS (A1) | PHENIX | 2021-02-02 | 35217718 | - | - | - | - | N/A | - | - | 3C-like proteinase | 13.7 | - | No | - | 8.9 | 36.8 | 53.7 | - | |||||||
| 7lmc | 2.98 Å | 2021-04-14 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Protein-protein complex | 20.76% | 28.81% | N/A | - | XDS | 88.70% | 10.6 | APS (17-ID) | BUSTER | 2021-02-05 | - | - | - | - | - | N/A | - | - | 3C-like proteinase; Non-structural protein 4 peptide | 13.7 | - | No | - | - | 10.6 | 65.8 | 22.9 | - | ||||||
| 7lxw | 2.80 Å | 2021-04-14 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2021-03-05 | 33761326 | - | - | - | - | N/A | - | - | Spike glycoprotein; S2X333 Fab Heavy Chain variable region; S2X333 Fab Light Chain variable region | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7lxx | 3.00 Å | 2021-04-14 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-03-05 | 33761326 | - | - | - | - | N/A | - | - | Spike glycoprotein; S2L28 Fab Light Chain variable region; S2L28 Fab Heavy Chain variable region | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lxy | 2.20 Å | 2021-04-14 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-03-05 | 33761326 | - | - | - | - | N/A | - | - | S2X333 Fab Light Chain variable region; S2X333 Fab Heavy Chain variable region; S2M11 Fab Heavy Chain variable region; S2M11 Fab Light Chain variable region; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7lxz | 2.60 Å | 2021-04-14 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-03-05 | 33761326 | - | - | - | - | N/A | - | - | S2L28 Fab Light Chain variable region; S2L28 Fab Heavy Chain variable region; S2M11 Fab Heavy Chain variable region; S2M11 Fab Light Chain variable region; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7ly0 | 2.60 Å | 2021-04-14 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-03-05 | 33761326 | - | - | - | - | N/A | - | - | S2M28 Fab Heavy Chain variable region; S2M28 Fab Light Chain variable region; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7ly2 | 2.50 Å | 2021-04-14 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-03-05 | 33761326 | - | - | - | - | N/A | - | - | S2M28 Fab Light Chain variable region; S2M28 Fab Heavy Chain variable region; S2M11 Fab Heavy Chain variable region; S2M11 Fab Light Chain variable region; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ly3 | 3.00 Å | 2021-04-14 | X-ray | XYL, UNX, NAG | SARS-CoV-2 | Spike | Protein-protein complex | 20.85% | 23.39% | N/A | Xylitol; UNKNOWN ATOM OR ION; 2-acetamido-2-deoxy-beta-D-glucopyranose | Xylitol; UNKNOWN ATOM OR ION; 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 100.00% | 12.6 | ALS (5.0.2) | PHENIX | 2021-03-05 | 33761326 | - | - | - | - | N/A | - | - | S2M28 Fab Light Chain; S2M28 Fab Heavy Chain; Spike protein S1 | 72.7 | - | No | - | 51.3 | 91.1 | 79.7 | - | ||||||
| 7me0 | 2.48 Å | 2021-04-14 | Cryo-EM | - | SARS-CoV-2 | NSP15 | No functional ligands | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-04-06 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7d7k | 1.90 Å | 2021-04-21 | X-ray | CFF | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 18.28% | 20.74% | 12.80% | CAFFEINE | CAFFEINE | XDS | 98.80% | 25.84 | SSRF (BL17U1) | PHENIX | 2020-10-04 | 33864621 | ZN | - | - | - | N/A | - | - | Non-structural protein 3 | 52.8 | - | No | - | 76.2 | 43.4 | 60.9 | - | ||||||
| 7d7l | 2.11 Å | 2021-04-21 | X-ray | GXU, CFF | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 18.33% | 21.83% | 28.10% | 1-(2-methoxyethyl)-2-methyl-3-(pyrazin-2-ylmethyl)benzo[f]benzimidazol-3-ium-4,9-dione; CAFFEINE | 1-(2-methoxyethyl)-2-methyl-3-(pyrazin-2-ylmethyl)benzo[f]benzimidazol-3-ium-4,9-dione; CAFFEINE | XDS | 99.90% | 17.71 | SLS (X06SA) | PHENIX | 2020-10-04 | 33864621 | ZN | - | - | - | N/A | - | - | Non-structural protein 3 | 47.9 | - | No | - | 66.7 | 34.8 | 69.6 | - | ||||||
| 7dx4 | 3.60 Å | 2021-04-21 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-01-18 | 34676098 | - | - | - | - | N/A | - | - | Light chain of FC08 Fab; Heavy chain of FC08 Fab; Spike protein S1; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7krn | 3.40 Å | 2021-04-21 | Cryo-EM | ADP, AF3, 1N7 | SARS-CoV-2 | NSP7/NSP8/NSP12/NSP13 | Protein-protein complex | N/A | N/A | N/A | ADENOSINE-5'-DIPHOSPHATE; RNA (43-MER); RNA (37-MER); ALUMINUM FLUORIDE; CHAPSO | ADENOSINE-5'-DIPHOSPHATE; ALUMINUM FLUORIDE; CHAPSO | - | - | () | PHENIX | 2020-11-20 | 33883267 | MG; ZN | - | - | - | N/A | - | - | RNA (43-MER); RNA (37-MER); Helicase - UNP residues 5325-5925; Non-structural protein 7 - UNP residues 3860-3942; Non-structural protein 8 - UNP residues 3943-4140; RNA-directed RNA polymerase - UNP residues 4393-5324 | N/A | - | No | - | N/A | N/A | N/A | RNA (43-MER); RNA (37-MER) | - | ||||||
| 7kro | 3.60 Å | 2021-04-21 | Cryo-EM | ADP, AF3, 1N7 | SARS-CoV-2 | NSP7/NSP8/NSP12/NSP13 | Protein-protein complex | N/A | N/A | N/A | ADENOSINE-5'-DIPHOSPHATE; RNA (43-MER); RNA (37-MER); ALUMINUM FLUORIDE; CHAPSO | ADENOSINE-5'-DIPHOSPHATE; ALUMINUM FLUORIDE; CHAPSO | - | - | () | PHENIX | 2020-11-20 | 33883267 | MG; ZN | - | - | - | N/A | - | - | RNA (43-MER); RNA (37-MER); Helicase - UNP residues 5325-5925; Non-structural protein 7 - UNP residues 3860-3942; Non-structural protein 8 - UNP residues 3943-4140; RNA-directed RNA polymerase - UNP residues 4393-5324 | N/A | - | No | - | N/A | N/A | N/A | RNA (37-MER); RNA (43-MER) | - | ||||||
| 7krp | 3.20 Å | 2021-04-21 | Cryo-EM | 1N7, ADP | SARS-CoV-2 | NSP7/NSP8/NSP12 | Protein-protein complex | N/A | N/A | N/A | CHAPSO; ADENOSINE-5'-DIPHOSPHATE; RNA (36-MER); RNA (37-MER) | CHAPSO; ADENOSINE-5'-DIPHOSPHATE | - | - | () | PHENIX | 2020-11-20 | 33883267 | MG; ZN | - | - | - | N/A | - | - | RNA (36-MER); RNA (37-MER); Non-structural protein 7 - UNP residues 3860-3942; Non-structural protein 8 - UNP residues 3943-4140; RNA-directed RNA polymerase - UNP residues 4393-5324 | N/A | - | No | - | N/A | N/A | N/A | RNA (37-MER); RNA (36-MER) | - | ||||||
| 7b14 | 3.79 Å | 2021-04-28 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-11-23 | 33436526 | - | - | - | - | N/A | - | - | Nanobody against SARS-CoV-2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7b18 | 2.62 Å | 2021-04-28 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-11-24 | 33436526 | - | - | - | - | N/A | - | - | Nanobody against spike glycoprotein VHH V; Nanobody against SARS-CoV-2 VHH E; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7b62 | 1.82 Å | 2021-04-28 | X-ray | 1PE, PGE, PG4, NAG, BLA | SARS-CoV-2 | Spike | Protein-protein complex | 16.39% | 19.98% | 16.60% | PENTAETHYLENE GLYCOL; TRIETHYLENE GLYCOL; 2-acetamido-2-deoxy-beta-D-glucopyranose; BILIVERDINE IX ALPHA | PENTAETHYLENE GLYCOL; TRIETHYLENE GLYCOL; TETRAETHYLENE GLYCOL; 2-acetamido-2-deoxy-beta-D-glucopyranose; BILIVERDINE IX ALPHA | XDS | 99.40% | 12.4 | SLS (X06SA) | PHENIX; PHENIX | 2020-12-07 | 33888467 | - | - | - | - | N/A | - | - | Spike glycoprotein | 68.4 | - | No | - | 81.9 | 64.4 | 66.0 | - | ||||||
| 7nt9 | 3.36 Å | 2021-04-28 | Cryo-EM | NAG, BLA | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; BILIVERDINE IX ALPHA | 2-acetamido-2-deoxy-beta-D-glucopyranose; BILIVERDINE IX ALPHA | - | - | () | PHENIX | 2021-03-09 | 33888467 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7nta | 3.50 Å | 2021-04-28 | Cryo-EM | NAG, BLA | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; BILIVERDINE IX ALPHA | 2-acetamido-2-deoxy-beta-D-glucopyranose; BILIVERDINE IX ALPHA | - | - | () | PHENIX | 2021-03-09 | 33888467 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7ntc | 3.60 Å | 2021-04-28 | Cryo-EM | NAG, BLA | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; BILIVERDINE IX ALPHA; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose; BILIVERDINE IX ALPHA | - | - | () | PHENIX | 2021-03-09 | 33888467 | - | - | - | - | N/A | - | - | P008_056 Fab Light chain; P008_056 Fab Heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 6ln0 | 2.46 Å | 2021-05-05 | X-ray | - | SARS-CoV-2 | NSP3 | No functional ligands | 20.81% | 25.94% | 14.00% | - | HKL-2000 | 98.80% | 8.0 | SSRF (BL17U) | PHENIX | 2019-12-27 | 33327866 | ZN | - | - | - | N/A | - | - | nonstructural protein 3 | 41.3 | - | No | - | 57.0 | 73.5 | 27.9 | - | |||||||
| 7cam | 2.85 Å | 2021-05-05 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 20.13% | 30.92% | 14.90% | - | HKL-2000 | 95.20% | 10.6 | NSRRC (BL15A1) | PHENIX | 2020-06-09 | 32905393 | - | - | - | - | N/A | - | - | 3C-like proteinase | 29.1 | - | No | - | 6.0 | 93.1 | 35.1 | - | |||||||
| 7cb7 | 1.69 Å | 2021-05-05 | X-ray | B1S, K36, NA | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 15.66% | 19.16% | 2.30% | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; SODIUM ION | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; SODIUM ION | HKL-2000 | 97.61% | 24.3 | NSRRC (BL15A1) | PHENIX | 2020-06-10 | 32905393 | - | - | - | - | N/A | - | - | 3C-like proteinase | 64.2 | - | No | - | 86.9 | 50.6 | 65.8 | - | ||||||
| 7e23 | 3.30 Å | 2021-05-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-02-04 | 33893388 | - | - | - | - | N/A | - | - | CA521 Light Chain; CA521 Heavy Chain; Spike protein S1 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7ldj | 2.36 Å | 2021-05-05 | X-ray | PG4, NAG, MAN | SARS-CoV-2 | Spike | Functional ligand | 21.03% | 26.81% | 8.40% | alpha-D-mannopyranose | TETRAETHYLENE GLYCOL; 2-acetamido-2-deoxy-beta-D-glucopyranose; alpha-D-mannopyranose | XDS | 100.00% | 12.8 | Australian Synchrotron (MX2) | PHENIX | 2021-01-13 | - | - | - | - | - | N/A | - | - | Nanobody 2; Spike glycoprotein - receptor binding domain (UNP residues 331-527) | 21.6 | - | No | - | - | 20.7 | 44.6 | 52.8 | - | |||||
| 7lw3 | 2.30 Å | 2021-05-05 | X-ray | YG4, SAH, MES | SARS-CoV-2 | NSP10/NSP16 | Functional ligand | 22.24% | 25.11% | 8.90% | [(2~{R},3~{R},4~{R},5~{R})-5-(6-azanyl-7,8-dihydropurin-9-yl)-2-[[[[[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1,8-dihydropurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-4-methoxy-oxolan-3-yl] [(2~{R},3~{S},4~{R},5~{S})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate; S-ADENOSYL-L-HOMOCYSTEINE; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | [(2~{R},3~{R},4~{R},5~{R})-5-(6-azanyl-7,8-dihydropurin-9-yl)-2-[[[[[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1,8-dihydropurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-4-methoxy-oxolan-3-yl] [(2~{R},3~{S},4~{R},5~{S})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate; S-ADENOSYL-L-HOMOCYSTEINE; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | XDS | 99.90% | 15.2 | APS (24-ID-C) | PHENIX | 2021-02-27 | 34078893 | ZN; MG | - | - | - | N/A | - | - | Non-structural protein 10; 2'-O-methyltransferase | 18.3 | - | No | - | 34.1 | 21.8 | 54.8 | - | ||||||
| 7lw4 | 2.50 Å | 2021-05-05 | X-ray | SAH, MES | SARS-CoV-2 | NSP10/NSP16 | Functional ligand | 20.38% | 24.05% | 11.30% | S-ADENOSYL-L-HOMOCYSTEINE; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | S-ADENOSYL-L-HOMOCYSTEINE; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | XDS | 99.90% | 8.7 | APS (24-ID-C) | PHENIX | 2021-02-27 | 34078893 | ZN | - | - | - | N/A | - | - | Non-structural protein 10; 2'-O-methyltransferase | 57.3 | - | No | - | 44.4 | 59.4 | 85.3 | - | ||||||
| 7lx5 | 3.44 Å | 2021-05-05 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | 2021-03-03 | 33893175 | - | - | - | - | N/A | - | - | WNb 10; WNb 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7m6d | 3.10 Å | 2021-05-05 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 21.85% | 25.91% | 27.10% | - | XDS | 97.90% | 4.0 | SSRL (BL12-2) | PHENIX | 2021-03-25 | 34015271 | - | - | - | - | N/A | - | - | BG4-25 Fab Light Chain; BG4-25 Fab Heavy Chain; Spike protein S1 - Receptor Binding Domain; CR3022 Fab Light Chain; CR3022 Fab Heavy Chain | 20.2 | - | No | - | 27.2 | 51.2 | 36.4 | - | |||||||
| 7m6e | 3.30 Å | 2021-05-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-03-25 | 34015271 | - | - | - | - | N/A | - | - | BG10-19 Fab Light Chain; BG10-19 Fab Heavy Chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7m6f | 3.90 Å | 2021-05-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-03-25 | 34015271 | - | - | - | - | N/A | - | - | BG1-22 Fab Light Chain; BG1-22 Fab Heavy Chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7m6g | 3.70 Å | 2021-05-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-03-25 | 34015271 | - | - | - | - | N/A | - | - | BG7-15 Fab Light Chain; BG7-15 Fab Heavy Chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7m6h | 4.00 Å | 2021-05-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-03-25 | 34015271 | - | - | - | - | N/A | - | - | BG7-20 Fab Light Chain; BG7-20 Fab Heavy Chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7m6i | 4.00 Å | 2021-05-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-03-25 | 34015271 | - | - | - | - | N/A | - | - | BG1-24 Fab Light Chain; BG1-24 Fab Heavy Chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7m7w | 2.65 Å | 2021-05-05 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 22.12% | 27.10% | 14.90% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 98.60% | 10.9 | SSRL (BL9-2) | REFMAC | 2021-03-29 | 34261126 | - | - | - | - | N/A | - | - | Spike protein S1 - receptor binding domain (UNP residues 328-531); Monoclonal antibody S2H97 Fab heavy chain; Monoclonal antibody S2H97 Fab light chain; Monoclonal antibody S2X259 Fab heavy chain; Monoclonal antibody S2X259 Fab light chain | 38.3 | - | No | - | 18.9 | 66.8 | 66.9 | - | ||||||
| 7m8k | - | 2021-05-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-03-29 | 33887205 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7mc5 | 1.64 Å | 2021-05-05 | X-ray | TLA | SARS-CoV-2 | NSP10/NSP14 | Protein-protein complex | 16.63% | 19.69% | 11.40% | L(+)-TARTARIC ACID | L(+)-TARTARIC ACID | XDS | 98.90% | 12.2 | APS (24-ID-C) | PHENIX | 2021-04-01 | 35165203 | ZN | - | - | - | N/A | - | - | Non-structural protein 10 - UNP residues 4254-4392; Proofreading exoribonuclease - UNP residues 5926-6214 | 81.8 | - | No | - | 83.8 | 91.2 | 74.3 | - | ||||||
| 7mc6 | 2.10 Å | 2021-05-05 | X-ray | - | SARS-CoV-2 | NSP10/NSP14 | Protein-protein complex | 19.71% | 21.87% | 6.00% | - | XDS | 97.80% | 11.2 | APS (24-ID-C) | PHENIX | 2021-04-01 | 35165203 | MG; ZN | - | - | - | N/A | - | - | Non-structural protein 10 - UNP residues 4254-4392; Proofreading exoribonuclease - UNP residues 5926-6214 | 70.1 | - | No | - | 66.4 | 53.9 | 95.7 | - | |||||||
| 7b27 | 2.90 Å | 2021-05-12 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 26.61% | 30.51% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 93.50% | 7.8 | SLS (X06SA) | REFMAC | 2020-11-26 | 33904225 | - | - | - | - | N/A | - | - | neutralizing nanobody NM1230; Surface glycoprotein | 17.9 | - | No | - | 6.5 | 74.4 | 28.6 | - | ||||||
| 7dhg | 2.20 Å | 2021-05-12 | X-ray | - | SARS-CoV-2 | ORF9B | Protein-protein complex | 22.41% | 26.68% | 9.90% | - | XDS | 98.10% | 10.33 | SSRF (BL17U) | PHENIX | 2020-11-14 | 33990585 | - | - | - | - | N/A | - | - | ORF9b protein; Mitochondrial import receptor subunit TOM70 | 45.3 | - | No | - | 21.8 | 83.5 | 60.8 | - | |||||||
| 7dpp | 2.10 Å | 2021-05-12 | X-ray | MYC | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.49% | 22.02% | 9.10% | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE | HKL-3000 | 99.70% | 4.1 | SSRF (BL18U1) | PHENIX | 2020-12-21 | 34131140 | - | - | - | - | N/A | - | - | 3C-like proteinase | 72.6 | - | No | - | 64.8 | 63.1 | 93.9 | - | ||||||
| 7dpu | 1.75 Å | 2021-05-12 | X-ray | HER | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.47% | 19.90% | 11.20% | 7-methoxy-3,5-bis(oxidanyl)-2-[3,4,5-tris(oxidanyl)phenyl]chromen-4-one | 7-methoxy-3,5-bis(oxidanyl)-2-[3,4,5-tris(oxidanyl)phenyl]chromen-4-one | HKL-3000 | 97.70% | 4.9 | SSRF (BL18U1) | PHENIX | 2020-12-21 | 34131140 | - | - | - | - | N/A | - | - | 3C-like proteinase | 85.5 | - | No | - | 82.3 | 88.6 | 97.7 | - | ||||||
| 7dpv | 2.35 Å | 2021-05-12 | X-ray | HF0 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.21% | 23.36% | 28.30% | (2S,3S)-3,5-dihydroxy-7-methoxy-2-(3,4,5-trihydroxyphenyl)chroman-4-one | (2S,3S)-3,5-dihydroxy-7-methoxy-2-(3,4,5-trihydroxyphenyl)chroman-4-one | HKL-3000 | 99.50% | 3.5 | SSRF (BL18U1) | PHENIX | 2020-12-21 | 34131140 | - | - | - | - | N/A | - | - | 3C-like proteinase | 62.6 | - | No | - | 51.5 | 55.3 | 93.1 | - | ||||||
| 7lcp | 1.90 Å | 2021-05-12 | X-ray | CA, UED | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.53% | 22.08% | N/A | CALCIUM ION; N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | CALCIUM ION; N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | XDS | 98.04% | 16.12 | SSRL (BL12-2) | PHENIX | 2021-01-11 | 33895266 | CA | - | - | - | N/A | - | - | 3C-like proteinase | 30.1 | - | No | - | 64.4 | 39.8 | 31.9 | - | ||||||
| 7lcq | 2.15 Å | 2021-05-12 | X-ray | UED | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.55% | 22.32% | N/A | N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | XDS | 96.94% | 14.77 | SSRL (BL12-2) | PHENIX | 2021-01-11 | 33895266 | - | - | - | - | N/A | - | - | 3C-like proteinase | 22.2 | - | No | - | 61.9 | 25.0 | 32.5 | - | ||||||
| 7m3i | 2.80 Å | 2021-05-12 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 24.54% | 28.11% | 3.40% | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.48% | 12.1 | APS (19-ID) | PHENIX | 2021-03-18 | 34237283 | - | - | - | - | N/A | - | - | Spike protein S1 - receptor binding domain; CV2-75 Fab Light chain; CV2-75 Fab Heavy chain | 12.1 | - | No | - | 13.5 | 25.0 | 56.4 | - | |||||||
| 7mf1 | 2.09 Å | 2021-05-12 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 20.68% | 25.03% | 9.50% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 98.00% | 5.5 | NSLS-II (17-ID-2) | PHENIX | 2021-04-08 | 33932326 | - | - | - | - | N/A | - | - | 47D1 Fab light chain; 47D1 Fab heavy chain; Spike protein S1 - Receptor binding domain, UNP residues 333-530 | 43.7 | - | No | - | 34.9 | 69.8 | 58.3 | - | ||||||
| 7mhf | 1.55 Å | 2021-05-12 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 18.34% | 22.23% | 15.79% | - | xia2 | 99.72% | 3.2643 | NSLS-II (17-ID-2) | PHENIX | 2021-04-15 | 36071812 | - | - | - | - | N/A | - | - | 3C-like proteinase | 52.6 | - | No | - | 61.0 | 48.9 | 70.2 | - | |||||||
| 7mhg | 1.53 Å | 2021-05-12 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 17.01% | 20.43% | 18.01% | - | xia2 | 99.97% | 7.8732 | NSLS-II (17-ID-2) | PHENIX | 2021-04-15 | 36071812 | - | - | - | - | N/A | - | - | 3C-like proteinase | 74.3 | - | No | - | 78.0 | 64.4 | 83.5 | - | |||||||
| 7mhh | 2.19 Å | 2021-05-12 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 19.94% | 25.47% | 29.16% | - | xia2 | 99.89% | 4.4679 | NSLS-II (17-ID-2) | PHENIX | 2021-04-15 | 36071812 | - | - | - | - | N/A | - | - | 3C-like proteinase | 38.8 | - | No | - | 33.0 | 45.7 | 74.8 | - | |||||||
| 7mhi | 1.88 Å | 2021-05-12 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 18.57% | 22.76% | 18.18% | - | xia2 | 100.00% | 5.0456 | NSLS-II (17-ID-2) | PHENIX | 2021-04-15 | 36071812 | - | - | - | - | N/A | - | - | 3C-like proteinase | 61.3 | - | No | - | 57.5 | 48.9 | 90.6 | - | |||||||
| 7mhj | 2.00 Å | 2021-05-12 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 19.42% | 24.21% | 17.81% | - | xia2 | 99.03% | 6.2709 | NSLS-II (17-ID-2) | PHENIX | 2021-04-15 | 36071812 | - | - | - | - | N/A | - | - | 3C-like proteinase | 44.8 | - | No | - | 45.2 | 42.8 | 77.1 | - | |||||||
| 7mhk | 1.96 Å | 2021-05-12 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 20.39% | 24.82% | 19.52% | - | xia2 | 99.86% | 5.3964 | NSLS-II (17-ID-2) | PHENIX | 2021-04-15 | 36071812 | - | - | - | - | N/A | - | - | 3C-like proteinase | 38.6 | - | No | - | 37.8 | 42.8 | 72.7 | - | |||||||
| 7mhl | 1.55 Å | 2021-05-12 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 17.00% | 22.35% | 15.79% | - | xia2 | 99.72% | 3.2643 | NSLS-II (17-ID-2) | PHENIX | 2021-04-15 | 36071812 | - | - | - | - | N/A | - | - | 3C-like proteinase | 25.5 | - | No | - | 57.9 | 20.3 | 48.2 | - | |||||||
| 7mhm | 1.53 Å | 2021-05-12 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 16.06% | 19.78% | 18.01% | - | xia2 | 99.97% | 7.8732 | NSLS-II (17-ID-2) | PHENIX | 2021-04-15 | 36071812 | - | - | - | - | N/A | - | - | 3C-like proteinase | 45.0 | - | No | - | 83.9 | 37.2 | 44.5 | - | |||||||
| 7mhn | 2.19 Å | 2021-05-12 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 16.26% | 21.69% | 29.16% | - | xia2 | 99.89% | 4.4679 | NSLS-II (17-ID-2) | PHENIX | 2021-04-15 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 20.0 | - | No | - | - | 69.5 | 18.2 | 26.7 | - | ||||||
| 7mho | 1.88 Å | 2021-05-12 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 16.67% | 20.77% | 18.18% | - | xia2 | 100.00% | 5.0456 | NSLS-II (17-ID-2) | PHENIX | 2021-04-15 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 29.3 | - | No | - | - | 75.4 | 24.3 | 34.6 | - | ||||||
| 7mhp | 2.00 Å | 2021-05-12 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 15.92% | 21.74% | 17.81% | - | xia2 | 99.03% | 6.2709 | NSLS-II (17-ID-2) | PHENIX | 2021-04-15 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 17.4 | - | No | - | - | 63.8 | 19.3 | 25.5 | - | ||||||
| 7mhq | 1.96 Å | 2021-05-12 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 16.86% | 22.40% | 19.52% | - | xia2 | 99.86% | 5.3964 | NSLS-II (17-ID-2) | PHENIX | 2021-04-15 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 11.7 | - | No | - | - | 50.3 | 16.5 | 26.8 | - | ||||||
| 7mjg | 2.81 Å | 2021-05-12 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-04-20 | 33914735 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7mjh | 2.66 Å | 2021-05-12 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-04-20 | 33914735 | - | - | - | - | N/A | - | - | VH ab8; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7mji | 2.81 Å | 2021-05-12 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-04-20 | 33914735 | - | - | - | - | N/A | - | - | VH ab8; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7mjj | 3.32 Å | 2021-05-12 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-04-20 | 33914735 | - | - | - | - | N/A | - | - | Fab ab1 Light Chain; Fab ab1 Heavy Chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7mjk | 2.73 Å | 2021-05-12 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-04-20 | 33914735 | - | - | - | - | N/A | - | - | Fab ab1 Light Chain; Fab ab1 Heavy Chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7mjl | 2.95 Å | 2021-05-12 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-04-20 | 33914735 | - | - | - | - | N/A | - | - | Fab ab1 Light Chain; Fab ab1 Heavy Chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7mjm | 2.83 Å | 2021-05-12 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-04-20 | 33914735 | - | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7mjn | 3.29 Å | 2021-05-12 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-04-20 | 33914735 | - | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7mkl | 3.20 Å | 2021-05-12 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-04-24 | 34481543 | - | - | - | - | N/A | - | - | SARS2-38 Fv light chain; SARS2-38 Fv heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7mkm | 3.16 Å | 2021-05-12 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-04-24 | 34481543 | - | - | - | - | N/A | - | - | Spike protein S1 - receptor binding domain (UNP residues 333-520); SARS2-38 Fv light chain; SARS2-38 Fv heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7mmo | 2.43 Å | 2021-05-12 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 22.61% | 25.26% | 6.90% | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.80% | 15.0 | APS (31-ID) | BUSTER | 2021-04-30 | 33972947 | - | - | - | - | N/A | - | - | Spike protein S1 - receptor-binding domain; LY-CoV1404 Fab light chain; LY-CoV1404 Fab heavy chain | 46.2 | - | No | - | 32.9 | 41.1 | 93.8 | - | |||||||
| 7mng | 1.70 Å | 2021-05-12 | X-ray | ZL7 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.95% | 21.79% | 18.40% | (2R,3S)-N-cyclopropyl-3-{[(2R)-3-(cyclopropylmethanesulfonyl)-2-{[(1S)-2,2,2-trifluoro-1-(4-fluorophenyl)ethyl]amino}propanoyl]amino}-2-hydroxypentanamide (non-preferred name) | (2R,3S)-N-cyclopropyl-3-{[(2R)-3-(cyclopropylmethanesulfonyl)-2-{[(1S)-2,2,2-trifluoro-1-(4-fluorophenyl)ethyl]amino}propanoyl]amino}-2-hydroxypentanamide (non-preferred name) | XDS | 100.00% | 9.0 | NSLS-II (17-ID-2) | REFMAC | 2021-04-30 | 35842458 | - | - | - | - | N/A | - | - | 3C-like proteinase | 34.0 | - | No | - | 67.2 | 42.8 | 34.0 | - | ||||||
| 7mpb | 2.30 Å | 2021-05-12 | X-ray | ETF, ASC | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.99% | 24.40% | 20.00% | ASCORBIC ACID | TRIFLUOROETHANOL; ASCORBIC ACID | MOSFLM | 92.60% | 8.2 | APS (19-ID) | PHENIX | 2021-05-04 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 25.1 | - | No | - | - | 41.0 | 30.5 | 54.2 | - | |||||
| 7nkt | 2.30 Å | 2021-05-12 | X-ray | PO4, NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 18.43% | 22.39% | N/A | PHOSPHATE ION; 2-acetamido-2-deoxy-beta-D-glucopyranose | PHOSPHATE ION; 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.90% | 19.5 | SLS (X06DA) | REFMAC | 2021-02-18 | 33904225 | - | - | - | - | N/A | - | - | neutralizing nanobody NM1226; Spike protein S1 | 53.7 | - | No | - | 61.4 | 70.3 | 50.5 | - | ||||||
| 7ofs | 1.90 Å | 2021-05-12 | X-ray | YRL | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 19.39% | 22.43% | N/A | 4-(2-hydroxyethyl)phenol | 4-(2-hydroxyethyl)phenol | XDS | 99.92% | 15.25 | PETRA III, DESY (P11) | PHENIX; PHENIX | 2021-05-05 | 35953531 | ZN | - | - | - | N/A | - | - | Non-structural protein 3 | 50.8 | - | No | - | 70.4 | 16.3 | 89.8 | - | ||||||
| 7oft | 1.95 Å | 2021-05-12 | X-ray | HBA | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 18.29% | 21.35% | N/A | P-HYDROXYBENZALDEHYDE | P-HYDROXYBENZALDEHYDE | XDS | 99.95% | 20.61 | PETRA III, DESY (P11) | PHENIX; PHENIX | 2021-05-05 | 35953531 | ZN | - | - | - | N/A | - | - | Non-structural protein 3 | 52.2 | - | No | - | 72.8 | 16.7 | 89.7 | - | ||||||
| 7ofu | 1.72 Å | 2021-05-12 | X-ray | PO4, HE9 | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 17.92% | 20.87% | N/A | PHOSPHATE ION; methyl 3,4-bis(oxidanyl)benzoate | PHOSPHATE ION; methyl 3,4-bis(oxidanyl)benzoate | XDS | 100.00% | 16.66 | PETRA III, DESY (P11) | REFMAC | 2021-05-05 | 35953531 | ZN | - | - | - | N/A | - | - | Non-structural protein 3 | 66.4 | - | No | - | 80.3 | 57.1 | 70.4 | - | ||||||
| 7akd | 4.00 Å | 2021-05-19 | Cryo-EM | MAN, FUC, NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose; alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose; alpha-D-mannopyranose; alpha-L-fucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | alpha-D-mannopyranose; alpha-L-fucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-09-30 | 33958322 | - | - | - | - | N/A | - | - | 47D11 neutralizing antibody light chain; 47D11 neutralizing antibody heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7akj | 3.80 Å | 2021-05-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-10-01 | 33958322 | - | - | - | - | N/A | - | - | 47D11 neutralizing antibody light chain; 47D11 neutralizing antibody heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7c53 | 2.28 Å | 2021-05-19 | X-ray | CA | SARS-CoV-2 | Spike | Protein-protein complex | 19.51% | 24.18% | N/A | CALCIUM ION | CALCIUM ION | XDS | 99.70% | 17.12 | ROTATING ANODE () | PHENIX | 2020-05-19 | 34326308 | CA | - | - | - | N/A | - | - | Spike protein S2',pan-CoVs inhibitor EK1 - HR1 motif | 32.6 | - | No | - | 43.3 | 38.8 | 59.0 | - | ||||||
| 7cho | 2.56 Å | 2021-05-19 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 20.17% | 24.73% | 11.70% | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 95.90% | 9.5 | SSRF (BL17U) | PHENIX | 2020-07-06 | 34244522 | - | - | - | - | N/A | - | - | antibody P5A-1D2 light chain; antibody P5A-1D2 heavy chain; Spike protein S1 - receptor binding domain | 25.6 | - | No | - | 37.8 | 46.4 | 42.3 | - | |||||||
| 7chp | 2.36 Å | 2021-05-19 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 19.34% | 21.98% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 97.30% | 13.0 | SSRF (BL17U) | PHENIX | 2020-07-06 | 34244522 | - | - | - | - | N/A | - | - | antibody P5A-3C8 light chain; antibody P5A-3C8 heavy chain; Spike protein S1 - receptor binding domain | 50.6 | - | No | - | 65.4 | 42.0 | 68.9 | - | |||||||
| 7chs | 2.40 Å | 2021-05-19 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 19.08% | 22.44% | 11.40% | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 100.00% | 13.1 | SSRF (BL17U) | PHENIX | 2020-07-06 | 34244522 | - | - | - | - | N/A | - | - | antibody P22A-1D1 light chain; antibody P22A-1D1 heavy chain; Spike protein S1 - receptor binding domain | 46.4 | - | No | - | 60.8 | 62.2 | 45.2 | - | |||||||
| 7ddo | 3.40 Å | 2021-05-19 | Cryo-EM | NAG | Pangolin CoV | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-10-29 | 34018203 | ZN | - | - | - | N/A | - | - | Spike protein S1; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7ddp | 3.40 Å | 2021-05-19 | Cryo-EM | NAG | Pangolin CoV | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-10-29 | 34018203 | ZN | - | - | - | N/A | - | - | Spike protein S1; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7diy | 2.69 Å | 2021-05-19 | X-ray | - | SARS-CoV-2 | NSP10/NSP14 | Protein-protein complex | 23.01% | 26.37% | 12.80% | - | HKL-2000 | 99.60% | 11.0 | SSRF (BL18U1) | PHENIX | 2020-11-19 | 33956156 | MG; ZN | - | - | - | N/A | - | - | nsp14-ExoN protein - UNP residues 5926-6214; nsp10 protein - UNP residues 4254-4392 | 24.8 | - | No | - | 24.0 | 66.2 | 34.7 | - | |||||||
| 7m8j | 3.48 Å | 2021-05-19 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | 2021-03-29 | 33947773 | - | - | - | - | N/A | - | - | CM25 Fab - Light Chain; CM25 Fab - Heavy Chain; Spike protein S1 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7mrr | 2.32 Å | 2021-05-19 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Protein-protein complex | 19.40% | 24.55% | 11.50% | LEUPEPTIN | - | XDS | 98.30% | 12.1 | NSLS-II (17-ID-2) | REFMAC | 2021-05-08 | 35842458 | - | - | - | - | N/A | - | - | LEUPEPTIN; 3C-like proteinase | 54.9 | - | No | - | 39.7 | 64.7 | 80.1 | - | ||||||
| 7e8m | 2.09 Å | 2021-05-26 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 16.89% | 19.86% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 90.92% | 17.2 | SSRF (BL17U1) | PHENIX | 2021-03-02 | 34166623 | - | - | - | - | N/A | - | - | antibody P2C-1F11 light chain; antibody P2C-1F11 heavy chain; Spike protein S1 | 57.1 | - | No | - | 82.7 | 29.8 | 76.4 | - | ||||||
| 7kqb | 2.42 Å | 2021-05-26 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-11-14 | 33974910 | - | - | - | - | N/A | - | - | Fab 5A6 light chain; Fab 5A6 heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7kqe | 2.88 Å | 2021-05-26 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | 2020-11-15 | 33974910 | - | - | - | - | N/A | - | - | Spike glycoprotein; Fab 3D11 light chain; Fab 3D11 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7m5e | 2.50 Å | 2021-05-26 | Cryo-EM | SIA, NAG, FOL | MERS-CoV | Spike | Functional ligand | N/A | N/A | N/A | FOLIC ACID | N-acetyl-alpha-neuraminic acid; 2-acetamido-2-deoxy-beta-D-glucopyranose; FOLIC ACID | - | - | () | 2021-03-23 | 33981021 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7m71 | 2.66 Å | 2021-05-26 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-03-26 | 33974910 | - | - | - | - | N/A | - | - | Antibody 5A6 Fab light chain; Antibody 5A6 Fab heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7m7b | 2.95 Å | 2021-05-26 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-03-27 | 33974910 | - | - | - | - | N/A | - | - | Antibody Fab 3D11 light chain; Antibody Fab 3D11 heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7msw | 3.76 Å | 2021-05-26 | Cryo-EM | - | SARS-CoV-2 | NSP2 | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2021-05-12 | - | ZN | - | - | - | N/A | - | - | Non-structural protein 2 | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7msx | 3.15 Å | 2021-05-26 | Cryo-EM | - | SARS-CoV-2 | NSP2 | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2021-05-12 | - | ZN | - | - | - | N/A | - | - | Non-structural protein 2 | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7baj | 1.65 Å | 2021-06-02 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 18.55% | 20.66% | 3.80% | - | DIALS | 99.90% | 13.2 | Diamond (I04) | REFMAC | 2020-12-15 | 34031399 | - | - | - | - | N/A | - | - | Main Protease | 74.4 | - | No | - | 76.9 | 68.9 | 80.6 | - | |||||||
| 7bak | 2.05 Å | 2021-06-02 | X-ray | SE | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.00% | 21.93% | 4.80% | SELENIUM ATOM | SELENIUM ATOM | DIALS | 99.50% | 12.4 | Diamond (I04) | REFMAC | 2020-12-15 | 34031399 | - | - | - | - | N/A | - | - | Main Protease | 67.6 | - | No | - | 65.7 | 73.6 | 71.1 | - | ||||||
| 7bal | 1.85 Å | 2021-06-02 | X-ray | SE | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 22.39% | 24.90% | 7.50% | SELENIUM ATOM | SELENIUM ATOM | DIALS | 99.90% | 8.6 | Diamond (I04) | REFMAC | 2020-12-15 | 34031399 | - | - | - | - | N/A | - | - | Main Protease | 50.9 | - | No | - | 36.0 | 73.2 | 67.5 | - | ||||||
| 7dzw | 3.45 Å | 2021-06-02 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2021-01-26 | 34139176 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7dzx | 3.53 Å | 2021-06-02 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | 2021-01-26 | 34139176 | - | - | - | - | N/A | - | - | Fab Heavy chain of enhancing antibody; Fab light chain of enhancing antibody; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7dzy | 3.60 Å | 2021-06-02 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | 2021-01-26 | 34139176 | - | - | - | - | N/A | - | - | Fab light chain of enhancing antibody 2490; Spike glycoprotein; Fab Heavy chain of enhancing antibody 2490 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7mfu | 1.70 Å | 2021-06-02 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 18.11% | 21.49% | 8.60% | - | XDS | 98.44% | 8.87 | APS (22-ID) | PHENIX | 2021-04-11 | 34537245 | - | - | - | - | N/A | - | - | Synthetic nanobody #68 (Sb68); Synthetic Nanobody #14 (Sb14); Spike protein S1 - Receptor Binding Domain (RBD) | 73.7 | - | No | - | 70.0 | 64.6 | 89.9 | - | |||||||
| 7n06 | 2.20 Å | 2021-06-02 | Cryo-EM | - | SARS-CoV-2 | NSP15 | No functional ligands | N/A | N/A | N/A | RNA (5'-R(*AP*UP*A)-3') | - | - | - | () | phenix.real_space_refine; PHENIX | 2021-05-25 | 34403466 | - | - | - | - | N/A | - | - | RNA (5'-R(*AP*UP*A)-3'); Uridylate-specific endoribonuclease | N/A | - | No | - | N/A | N/A | N/A | RNA (5'-R(*AP*UP*A)-3') | - | ||||||
| 7n0g | 3.02 Å | 2021-06-02 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-05-25 | 34537245 | - | - | - | - | N/A | - | - | Spike glycoprotein; Synthetic nanobody (sybody), Sb45 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7n0h | 3.34 Å | 2021-06-02 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-05-25 | 34537245 | - | - | - | - | N/A | - | - | Synthetic nanobody (Sb45); Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7n0r | 1.42 Å | 2021-06-02 | X-ray | - | SARS-CoV-2 | Nucleocapsid | Pathogen-host interaction | 14.05% | 16.62% | 3.70% | - | XDS | 99.10% | 25.7 | APS (24-ID-E) | PHENIX | 2021-05-25 | 34381460 | - | - | - | - | N/A | - | - | Single-domain antibody C2; Nucleoprotein - RNA-binding domain | 77.5 | - | No | - | 96.0 | 55.4 | 83.4 | - | |||||||
| 7o7y | 2.20 Å | 2021-06-02 | Cryo-EM | SPD, UNX, SPM | SARS-CoV-2 | Ribosome | Functional ligand | N/A | N/A | N/A | mRNA containing SARS-CoV-2 sequence; UNKNOWN ATOM OR ION; SPERMIDINE; SPERMINE | SPERMIDINE; UNKNOWN ATOM OR ION; SPERMINE | - | - | () | 2021-04-14 | 34029205 | MG; ZN | - | - | - | N/A | - | - | 60S ribosomal protein L9; 18S rRNA; E-site tRNA; P-site Phe-tRNA(Phe); 40S ribosomal protein SA; 40S ribosomal protein S3a; Ribosomal protein uS5; 40S ribosomal protein S3; Ribosomal protein eS4; Ribosomal protein S5; 40S ribosomal protein S6; 40S ribosomal protein S7; 40S ribosomal protein S27; 40S ribosomal protein S8; Ribosomal protein S9 (Predicted); eS10; 40S ribosomal protein S11; 40S ribosomal protein S12; uS15; 40S ribosomal protein uS11; 40S ribosomal protein uS19; uS9; 40S ribosomal protein eS17; Ribosomal protein S28; 40S ribosomal protein S18; Ribosomal protein eS19; 40S ribosomal protein uS10; Ribosomal protein eS21; Ribosomal protein S15a; 40S ribosomal protein S23; 40S ribosomal protein S24; 40S ribosomal protein S25; 60s ribosomal protein l41; 28S rRNA; Ribosomal protein S27a; 5S rRNA; 5.8S rRNA; Ribosomal protein uL2; Ribosomal protein L3; 60S ribosomal protein L4; Ribosomal_L18_c domain-containing protein; 60S ribosomal protein L6; Ribosomal Protein uL30; Ribosomal protein eL8; 40S ribosomal protein S30; 60S ribosomal protein L10; Ribosomal protein L11; Replicase polyprotein 1ab; Ribosomal protein eL13; Ribosomal protein L14; Ribosomal protein L15; Ribosomal protein uL13; uL22; Ribosomal Protein eL18; 60S ribosomal protein L19; Ribosomal protein eS26; Ribosomal protein eL20; eL21; Ribosomal protein eL22; Ribosomal protein L23; eL24; uL23; Ribosomal protein L26; 60S ribosomal protein L27; 60S ribosomal protein L27a; 60S ribosomal protein L29; RACK1; eL30; eL31; eL32; eL33; 60S ribosomal protein L34; uL29; 60S ribosomal protein L36; Ribosomal protein L37; eL38; eL39; uS14; 60S ribosomal protein L40; eL42; eL43; Ribosomal protein eL28; 60S acidic ribosomal protein P0; Ribosomal protein L12; Ribosomal protein uL1; mRNA containing SARS-CoV-2 sequence | N/A | - | No | - | N/A | N/A | N/A | mRNA containing SARS-CoV-2 sequence | - | |||||||
| 7o7z | 2.40 Å | 2021-06-02 | Cryo-EM | SPD, UNX, SPM | SARS-CoV-2 | Ribosome | Functional ligand | N/A | N/A | N/A | mRNA containing SARS-CoV-2 sequence; SPERMINE; UNKNOWN ATOM OR ION; SPERMIDINE | SPERMIDINE; UNKNOWN ATOM OR ION; SPERMINE | - | - | () | 2021-04-14 | 34029205 | MG; ZN | - | - | - | N/A | - | - | eL24; uL23; Ribosomal protein L26; 60S ribosomal protein L27; 60S ribosomal protein L27a; 60S ribosomal protein L29; RACK1; eL30; eL31; eL32; eL33; 60S ribosomal protein L34; uL29; 60S ribosomal protein L36; Ribosomal protein L37; eL38; eL39; uS14; 60S ribosomal protein L40; eL42; eL43; Ribosomal protein eL28; 60S acidic ribosomal protein P0; Ribosomal protein L12; Ribosomal protein uL1; mRNA containing SARS-CoV-2 sequence; 18S rRNA; E-site tRNA; P-site Phe-tRNA(Phe); 40S ribosomal protein SA; 40S ribosomal protein S3a; Ribosomal protein uS5; 40S ribosomal protein S3; Ribosomal protein eS4; Ribosomal protein S5; 40S ribosomal protein S6; 40S ribosomal protein S7; 40S ribosomal protein S27; 40S ribosomal protein S8; Ribosomal protein S9 (Predicted); eS10; 40S ribosomal protein S11; 40S ribosomal protein S12; uS15; 40S ribosomal protein uS11; 40S ribosomal protein uS19; uS9; 40S ribosomal protein eS17; Ribosomal protein S28; 40S ribosomal protein S18; Ribosomal protein eS19; 40S ribosomal protein uS10; Ribosomal protein eS21; Ribosomal protein S15a; 40S ribosomal protein S23; 40S ribosomal protein S24; 40S ribosomal protein S25; 60s ribosomal protein l41; 28S rRNA; Ribosomal protein S27a; 5S; 5.8S rRNA; Ribosomal protein uL2; Ribosomal protein L3; 60S ribosomal protein L4; Ribosomal_L18_c domain-containing protein; 60S ribosomal protein L6; Ribosomal Protein uL30; Ribosomal protein eL8; 60S ribosomal protein L9; 40S ribosomal protein S30; 60S ribosomal protein L10; Ribosomal protein L11; Replicase polyprotein 1ab; Ribosomal protein eL13; Ribosomal protein L14; Ribosomal protein L15; Ribosomal protein uL13; uL22; Ribosomal Protein eL18; 60S ribosomal protein L19; Ribosomal protein eS26; Ribosomal protein eL20; eL21; Ribosomal protein eL22; Ribosomal protein L23 | N/A | - | No | - | N/A | N/A | N/A | mRNA containing SARS-CoV-2 sequence | - | |||||||
| 7o80 | 2.90 Å | 2021-06-02 | Cryo-EM | SPD, SF4, GTP, UNX | SARS-CoV-2 | Ribosome | Functional ligand | N/A | N/A | N/A | IRON/SULFUR CLUSTER; SPERMIDINE; mRNA containing SARS-CoV-2 sequence; UNKNOWN ATOM OR ION; GUANOSINE-5'-TRIPHOSPHATE | SPERMIDINE; IRON/SULFUR CLUSTER; GUANOSINE-5'-TRIPHOSPHATE; UNKNOWN ATOM OR ION | - | - | () | 2021-04-14 | 34029205 | MG; ZN | - | - | - | N/A | - | - | E-site tRNA; 60S ribosomal protein L27; 60S ribosomal protein L27a; 60S ribosomal protein L29; RACK1; eL30; eL31; eL32; eL33; 60S ribosomal protein L34; uL29; 60S ribosomal protein L36; Ribosomal protein L37; eL38; eL39; uS14; 60S ribosomal protein L40; eL42; eL43; Ribosomal protein eL28; 60S acidic ribosomal protein P0; Ribosomal protein L12; Ribosomal protein; Eukaryotic peptide chain release factor subunit 1; ATP binding cassette subfamily E member 1; mRNA containing SARS-CoV-2 sequence; uL23; eL24; Ribosomal protein L23; Ribosomal protein eL22; eL21; Ribosomal protein L18a; Ribosomal protein eS26; 60S ribosomal protein L19; Ribosomal protein L18; uL22; Ribosomal protein L13a; Ribosomal protein L15; Ribosomal protein L14; Ribosomal protein eL13; Replicase polyprotein 1ab; Ribosomal protein L11; 60S ribosomal protein L10; 40S ribosomal protein S30; 60S ribosomal protein L9; Ribosomal protein eL8; Ribosomal Protein uL30; 60S ribosomal protein L6; Ribosomal_L18_c domain-containing protein; 60S ribosomal protein L4; Ribosomal protein L3; Ribosomal protein uL2; 5.8S rRNA; 5S rRNA; Ribosomal protein S27a; 28S rRNA; 60s ribosomal protein l41; 40S ribosomal protein S25; 40S ribosomal protein S24; 40S ribosomal protein S23; Ribosomal protein S15a; Ribosomal protein eS21; 40S ribosomal protein uS10; Ribosomal protein eS19; 40S ribosomal protein S18; Ribosomal protein S28; 40S ribosomal protein eS17; Ribosomal protein S16; 40S ribosomal protein uS19; 40S ribosomal protein uS11; uS15; 40S ribosomal protein S12; 40S ribosomal protein S11; Ribosomal protein eS10; Ribosomal protein S9 (Predicted); 40S ribosomal protein S8; 40S ribosomal protein S27; 40S ribosomal protein S7; 40S ribosomal protein S6; Ribosomal protein S5; Ribosomal protein eS4; 40S ribosomal protein S3; Ribosomal protein uS5; 40S ribosomal protein S3a; 40S ribosomal protein SA; P-site Leu-tRNA(Leu); Ribosomal protein L26; 18S rRNA | N/A | - | No | - | N/A | N/A | N/A | mRNA containing SARS-CoV-2 sequence | - | |||||||
| 7o81 | 3.10 Å | 2021-06-02 | Cryo-EM | UNX, SPD | SARS-CoV-2 | Ribosome | Functional ligand | N/A | N/A | N/A | mRNA containing SARS-CoV-2 sequence | UNKNOWN ATOM OR ION; SPERMIDINE | - | - | () | 2021-04-14 | 34029205 | MG; ZN | - | - | - | N/A | - | - | Ribosomal protein L23; Ribosomal protein uL1; Ribosomal protein L12; 60S acidic ribosomal protein P0; Ribosomal protein eL28; eL43; eL42; 60S ribosomal protein L40; eL39; uS14; eL38; Ribosomal protein L37; 60S ribosomal protein L36; uL29; 60S ribosomal protein L34; eL33; eL32; eL31; eL30; 60S ribosomal protein L29; RACK1; 60S ribosomal protein L27a; 60S ribosomal protein L27; Ribosomal protein L26; uL23; eL24; 40S ribosomal protein S11; Ribosomal protein eS10; Ribosomal protein S9 (Predicted); 40S ribosomal protein S8; 40S ribosomal protein S7; 40S ribosomal protein S27; 40S ribosomal protein S6; Ribosomal protein S5; Ribosomal protein eS4; 40S ribosomal protein S3; Ribosomal protein uS5; 40S ribosomal protein S3a; 40S ribosomal protein SA; EDF1; P-site tRNA(Pro); A-site Met-tRNA(Met); 18S rRNA; Ribosomal protein eL22; eL21; Ribosomal protein L18a; 60S ribosomal protein L19; Ribosomal protein eS26; Ribosomal protein L18; uL22; Ribosomal protein L13a; Ribosomal protein L15; Ribosomal protein L14; Ribosomal protein eL13; Replicase polyprotein 1ab; Ribosomal protein L11; 60S ribosomal protein L10; 60S ribosomal protein L9; mRNA containing SARS-CoV-2 sequence; 40S ribosomal protein S30; Ribosomal protein eL8; Ribosomal Protein uL30; 60S ribosomal protein L6; Ribosomal_L18_c domain-containing protein; 60S ribosomal protein L4; Ribosomal protein L3; Ribosomal protein uL2; 5.8S rRNA; 5S rRNA; 28S rRNA; Ribosomal protein S27a; 60s ribosomal protein l41; 40S ribosomal protein S25; 40S ribosomal protein S24; 40S ribosomal protein S23; Ribosomal protein S15a; Ribosomal protein eS21; 40S ribosomal protein uS10; Ribosomal protein eS19; 40S ribosomal protein S18; 40S ribosomal protein eS17; Ribosomal protein S28; uS9; 40S ribosomal protein uS19; 40S ribosomal protein uS11; uS15; 40S ribosomal protein S12 | N/A | - | No | - | N/A | N/A | N/A | mRNA containing SARS-CoV-2 sequence | - | |||||||
| 5sa4 | 2.05 Å | 2021-06-09 | X-ray | K3A | SARS-CoV-2 | NSP15 | PanDDA fragment screening | 20.40% | 22.52% | 19.70% | N-(5-methyl-1H-pyrazol-3-yl)acetamide | N-(5-methyl-1H-pyrazol-3-yl)acetamide | XDS | 99.60% | 6.4 | Diamond (I04-1) | BUSTER | 2021-05-19 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 67.5 | - | No | - | 60.0 | 70.3 | 80.0 | - | ||||||
| 5sa5 | 2.09 Å | 2021-06-09 | X-ray | ZQA | SARS-CoV-2 | NSP15 | PanDDA fragment screening | 17.86% | 21.91% | 25.70% | 4-ethyl-2-(1H-imidazol-5-yl)-1,3-thiazole | 4-ethyl-2-(1H-imidazol-5-yl)-1,3-thiazole | XDS | 100.00% | 5.4 | Diamond (I04-1) | BUSTER | 2021-05-19 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 69.7 | - | No | - | 65.8 | 85.6 | 63.7 | - | ||||||
| 5sa6 | 2.52 Å | 2021-06-09 | X-ray | O3G | SARS-CoV-2 | NSP15 | PanDDA fragment screening | 19.69% | 22.24% | 26.50% | N-benzyl-1-(4-fluorophenyl)methanamine | N-benzyl-1-(4-fluorophenyl)methanamine | XDS | 99.90% | 5.7 | Diamond (I04-1) | BUSTER | 2021-05-19 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 29.6 | - | No | - | 62.8 | 18.3 | 54.0 | - | ||||||
| 5sa7 | 2.22 Å | 2021-06-09 | X-ray | WL7 | SARS-CoV-2 | NSP15 | PanDDA fragment screening | 19.92% | 22.46% | 28.30% | 4-amino-N-(2-hydroxyethyl)-N-methylbenzene-1-sulfonamide | 4-amino-N-(2-hydroxyethyl)-N-methylbenzene-1-sulfonamide | XDS | 99.10% | 5.7 | Diamond (I04-1) | BUSTER | 2021-05-19 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 63.0 | - | No | - | 60.7 | 68.2 | 71.8 | - | ||||||
| 5sa8 | 2.30 Å | 2021-06-09 | X-ray | JOV | SARS-CoV-2 | NSP15 | PanDDA fragment screening | 19.09% | 22.24% | 35.90% | 3-chloro-N-(1-hydroxy-2-methylpropan-2-yl)benzamide | 3-chloro-N-(1-hydroxy-2-methylpropan-2-yl)benzamide | XDS | 100.00% | 5.8 | Diamond (I04-1) | BUSTER | 2021-05-19 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 75.7 | - | No | - | 62.8 | 85.6 | 81.5 | - | ||||||
| 5sa9 | 1.92 Å | 2021-06-09 | X-ray | GWG | SARS-CoV-2 | NSP15 | PanDDA fragment screening | 19.91% | 22.31% | 19.90% | 1-methylindazole-3-carboxamide | 1-methylindazole-3-carboxamide | XDS | 100.00% | 6.3 | Diamond (I04-1) | BUSTER | 2021-05-19 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 68.1 | - | No | - | 62.0 | 66.2 | 83.3 | - | ||||||
| 5saa | 2.24 Å | 2021-06-09 | X-ray | ZQD | SARS-CoV-2 | NSP15 | PanDDA fragment screening | 19.20% | 22.65% | 22.80% | 3-[(2S)-1-(methanesulfonyl)pyrrolidin-2-yl]-5-methyl-1,2-oxazole | 3-[(2S)-1-(methanesulfonyl)pyrrolidin-2-yl]-5-methyl-1,2-oxazole | XDS | 100.00% | 4.9 | Diamond (I04-1) | BUSTER | 2021-05-19 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 64.8 | - | No | - | 58.7 | 83.2 | 62.6 | - | ||||||
| 5sab | 2.49 Å | 2021-06-09 | X-ray | WJD | SARS-CoV-2 | NSP15 | PanDDA fragment screening | 18.56% | 23.07% | 54.10% | 2-methoxy-N-phenylacetamide | 2-methoxy-N-phenylacetamide | XDS | 99.50% | 4.1 | Diamond (I04-1) | BUSTER | 2021-05-19 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 60.8 | - | No | - | 54.4 | 91.9 | 49.7 | - | ||||||
| 5sac | 2.03 Å | 2021-06-09 | X-ray | VWG | SARS-CoV-2 | NSP15 | PanDDA fragment screening | 18.69% | 21.72% | 22.20% | N-hydroxyquinoline-2-carboxamide | N-hydroxyquinoline-2-carboxamide | XDS | 100.00% | 6.8 | Diamond (I04-1) | BUSTER | 2021-05-19 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 74.3 | - | No | - | 67.7 | 87.7 | 70.7 | - | ||||||
| 5sad | 1.96 Å | 2021-06-09 | X-ray | EJW | SARS-CoV-2 | NSP15 | PanDDA fragment screening | 19.70% | 22.27% | 18.00% | (3-phenyl-1,2-oxazol-5-yl)methylazanium | (3-phenyl-1,2-oxazol-5-yl)methylazanium | XDS | 99.90% | 7.0 | Diamond (I04-1) | BUSTER | 2021-05-19 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 61.5 | - | No | - | 62.5 | 56.9 | 78.2 | - | ||||||
| 5sae | 2.12 Å | 2021-06-09 | X-ray | W3G | SARS-CoV-2 | NSP15 | PanDDA fragment screening | 19.16% | 22.30% | 23.90% | pyridazin-3(2H)-one | pyridazin-3(2H)-one | XDS | 100.00% | 5.3 | Diamond (I04-1) | BUSTER | 2021-05-19 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 57.4 | - | No | - | 62.1 | 64.3 | 62.9 | - | ||||||
| 5saf | 2.11 Å | 2021-06-09 | X-ray | WOY | SARS-CoV-2 | NSP15 | PanDDA fragment screening | 19.22% | 22.19% | 31.60% | 6,7-dihydro-5H-pyrrolo[2,3-d]pyrimidine | 6,7-dihydro-5H-pyrrolo[2,3-d]pyrimidine | XDS | 100.00% | 6.3 | Diamond (I04-1) | BUSTER | 2021-05-19 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 72.1 | - | No | - | 63.4 | 83.2 | 74.1 | - | ||||||
| 5sag | 1.88 Å | 2021-06-09 | X-ray | ZQG | SARS-CoV-2 | NSP15 | PanDDA fragment screening | 20.41% | 23.00% | 14.30% | 3-(1H-imidazol-2-yl)propan-1-amine | 3-(1H-imidazol-2-yl)propan-1-amine | XDS | 100.00% | 7.8 | Diamond (I04-1) | BUSTER | 2021-05-19 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 69.4 | - | No | - | 54.9 | 68.2 | 91.3 | - | ||||||
| 5sah | 2.16 Å | 2021-06-09 | X-ray | ZQJ | SARS-CoV-2 | NSP15 | PanDDA fragment screening | 24.50% | 25.57% | 20.40% | 2-methyl-5,6,7,8-tetrahydropyrido[4,3-c]pyridazin-3(2H)-one | 2-methyl-5,6,7,8-tetrahydropyrido[4,3-c]pyridazin-3(2H)-one | XDS | 99.90% | 7.8 | Diamond (I04-1) | BUSTER | 2021-05-19 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 56.8 | - | No | - | 30.3 | 76.7 | 81.2 | - | ||||||
| 5sai | 2.02 Å | 2021-06-09 | X-ray | ZQM | SARS-CoV-2 | NSP15 | PanDDA fragment screening | 19.52% | 22.20% | 27.20% | N-{2-[(propan-2-yl)sulfanyl]phenyl}urea | N-{2-[(propan-2-yl)sulfanyl]phenyl}urea | XDS | 99.90% | 6.0 | Diamond (I04-1) | BUSTER | 2021-05-19 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 70.5 | - | No | - | 63.1 | 87.7 | 65.9 | - | ||||||
| 7cyh | 3.90 Å | 2021-06-09 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | PHENIX | 2020-09-03 | - | - | - | - | - | N/A | - | - | Heavy chain of HB27; Spike glycoprotein; Light chain of HB27 | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7cyp | 3.50 Å | 2021-06-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-09-04 | 34676096 | - | - | - | - | N/A | - | - | Heavy chain of HB27; SARS-CoV-2 Spike glycoprotein; Light chain of HB27 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7djz | 2.40 Å | 2021-06-09 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 18.32% | 22.98% | 14.90% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.70% | 4.8 | SSRF (BL19U1) | PHENIX | 2020-11-22 | 35332252 | - | - | - | - | N/A | - | - | MW01 light chain; MW01 heavy chain; Spike protein S1 - Receptor Binding Domain | 45.8 | - | No | - | 55.3 | 36.5 | 75.3 | - | ||||||
| 7dk0 | 3.20 Å | 2021-06-09 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 21.83% | 26.48% | 11.80% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 100.00% | 5.5 | SSRF (BL19U1) | PHENIX | 2020-11-22 | 35332252 | - | - | - | - | N/A | - | - | MW05 light chain; Spike protein S1 - Receptor Binding Domain; MW05 heavy chain | 48.2 | - | No | - | 23.2 | 93.4 | 55.1 | - | ||||||
| 7e7x | 2.78 Å | 2021-06-09 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 24.84% | 27.89% | N/A | - | HKL-2000 | 98.68% | 15.17 | SSRF (BL17U) | PHENIX | 2021-02-28 | 34021265 | - | - | - | - | N/A | - | - | N11 Fab heavy chain; Spike protein S1; N11 Fab Light chain | 11.5 | - | No | - | 14.7 | 25.3 | 53.0 | - | |||||||
| 7e7y | 2.41 Å | 2021-06-09 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 19.73% | 24.14% | N/A | - | HKL-2000 | 98.05% | 11.03 | Photon Factory (BL-1A) | PHENIX | 2021-02-28 | 34021265 | - | - | - | - | N/A | - | - | BD-623 Fab L; Spike protein S1; BD-623 Fab H | 56.7 | - | No | - | 43.5 | 78.9 | 65.5 | - | |||||||
| 7e86 | 2.90 Å | 2021-06-09 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 19.56% | 25.46% | 28.80% | - | HKL-2000 | 100.00% | 2.6 | NFPSS (BL19U1) | PHENIX | 2021-03-01 | 34021265 | - | - | - | - | N/A | - | - | Spike protein S1; BD-508 Fab Light Chain; BD-508 Fab Heavy Chain | 53.9 | - | No | - | 31.2 | 95.7 | 55.7 | - | |||||||
| 7e88 | 3.14 Å | 2021-06-09 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 21.73% | 25.48% | 7.20% | - | HKL-2000 | 97.70% | 5.7 | NFPSS (BL19U1) | PHENIX | 2021-03-01 | 34021265 | - | - | - | - | N/A | - | - | BD-515 Fab Light Chain; BD-515 Fab Heavy Chain; Spike protein S1 | 32.0 | - | No | - | 31.1 | 77.5 | 31.5 | - | |||||||
| 7e8c | 3.16 Å | 2021-06-09 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-03-01 | 34021265 | - | - | - | - | N/A | - | - | N9 L; 368-2 L; N9 H; 604 L; 604 H; Spike glycoprotein; 368-2 H | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7e8f | 3.18 Å | 2021-06-09 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-03-01 | 34021265 | - | - | - | - | N/A | - | - | N9 H; Spike protein S1; 368-2 L; 604 H; 604 L; Spike protein S1; 368-2 H; N9 L | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ek6 | 1.24 Å | 2021-06-09 | X-ray | - | Spike | Protein-protein complex | 17.31% | 18.54% | 5.80% | Spike protein S2; Spike protein S2 | - | XDS | 97.10% | 35.0 | SSRF (BL19U1) | PHENIX | 2021-04-04 | 34057039 | - | - | - | - | N/A | - | - | Spike protein S2; Spike protein S2 | 38.0 | - | No | - | 90.0 | 11.8 | 50.2 | - | |||||||
| 7eq4 | 1.25 Å | 2021-06-09 | X-ray | - | SARS-CoV-2 | NSP1 | Functional ligand | 16.33% | 19.72% | N/A | - | iMOSFLM | 99.86% | 36.33 | SSRF (BL17U1) | PHENIX; REFMAC | 2021-04-29 | 34132580 | - | - | - | - | N/A | - | - | Host translation inhibitor nsp1 | 65.5 | - | No | - | 83.5 | 29.7 | 92.8 | - | |||||||
| 7n0i | 2.20 Å | 2021-06-09 | X-ray | - | SARS-CoV-2 | Nucleocapsid | Pathogen-host interaction | 23.94% | 27.14% | 8.90% | - | XDS | 99.70% | 8.1 | ALS (5.0.2) | PHENIX | 2021-05-25 | 34381460 | MG | - | - | - | N/A | - | - | Nucleoprotein - C-terminal domain; Single-domain antibody E2 | 15.1 | - | No | - | 18.7 | 20.4 | 63.7 | - | |||||||
| 7n33 | 2.50 Å | 2021-06-09 | Cryo-EM | - | SARS-CoV-2 | NSP15 | No functional ligands | N/A | N/A | N/A | RNA (5'-R(*A)-D(*(UFT))-R(P*A)-3') | - | - | - | () | phenix.real_space_refine; PHENIX | 2021-05-31 | 34403466 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease; RNA (5'-R(*A)-D(*(UFT))-R(P*A)-3') | N/A | - | No | - | N/A | N/A | N/A | RNA (5'-R(*A)-D(*(UFT))-R(P*A)-3') | - | ||||||
| 7djr | 1.45 Å | 2021-06-16 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 16.64% | 20.10% | 7.70% | - | XDS | 96.10% | 15.3 | APS (21-ID-F) | PHENIX | 2020-11-21 | 34109016 | - | - | - | - | N/A | - | - | 3C-like proteinase | 66.8 | - | No | - | 81.0 | 56.0 | 71.7 | - | |||||||
| 7exm | 1.74 Å | 2021-06-16 | X-ray | - | SARS-CoV-2 | NSP2 | No functional ligands | 19.55% | 22.45% | N/A | - | HKL-3000 | 93.00% | 17.9 | SSRF (BL17U) | REFMAC | 2021-05-27 | 34398430 | ZN | - | - | - | N/A | - | - | Non-structural protein 2 | 53.7 | - | No | - | 59.7 | 45.8 | 76.8 | - | |||||||
| 7my2 | 2.65 Å | 2021-06-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-05-20 | 34098567 | - | - | - | - | N/A | - | - | Nanobody Nb30; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7my3 | 2.90 Å | 2021-06-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-05-20 | 34098567 | - | - | - | - | N/A | - | - | Nanobody Nb12; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7n5z | 1.76 Å | 2021-06-16 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 15.55% | 19.69% | 5.30% | - | XDS | 95.90% | 12.8 | LNLS SIRUS (MANACA) | PHENIX | 2021-06-07 | 34174328 | - | - | - | - | N/A | - | - | 3C-like proteinase | 82.2 | - | No | - | 83.8 | 88.7 | 78.2 | - | |||||||
| 7n6n | 2.80 Å | 2021-06-16 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Protein-protein complex | 19.92% | 25.54% | 27.90% | - | XDS | 99.60% | 3.3 | LNLS SIRUS (MANACA) | PHENIX | 2021-06-08 | 34174328 | - | - | - | - | N/A | - | - | 3C-like proteinase - UNP residues 3259-3569; 3C-like proteinase - N-terminal domain (UNP residues 3259-3263) | 36.1 | - | No | - | 30.4 | 78.9 | 39.1 | - | |||||||
| 7nt1 | 2.85 Å | 2021-06-16 | X-ray | UQW | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.41% | 27.84% | 18.10% | [(2R)-1-[2-(1H-indol-3-yl)ethylamino]-1-oxidanylidene-butan-2-yl] prop-2-enoate | [(2R)-1-[2-(1H-indol-3-yl)ethylamino]-1-oxidanylidene-butan-2-yl] prop-2-enoate | Aimless | 98.90% | 6.3 | SLS (X06SA) | REFMAC; REFMAC | 2021-03-08 | 34097796 | - | - | - | - | N/A | - | - | 3C-like proteinase | 35.6 | - | No | - | 14.9 | 86.1 | 46.2 | - | ||||||
| 7nt2 | 2.15 Å | 2021-06-16 | X-ray | URK | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.19% | 25.14% | 13.20% | [(1S)-2-[(2,3-dimethoxyphenyl)methylamino]-1-(4-nitrophenyl)-2-oxidanylidene-ethyl] prop-2-enoate | [(1S)-2-[(2,3-dimethoxyphenyl)methylamino]-1-(4-nitrophenyl)-2-oxidanylidene-ethyl] prop-2-enoate | Aimless | 98.10% | 9.8 | SLS (X06SA) | REFMAC; REFMAC | 2021-03-08 | 34097796 | - | - | - | - | N/A | - | - | 3C-like proteinase | 34.6 | - | No | - | 33.9 | 59.8 | 51.5 | - | ||||||
| 7nt3 | 2.33 Å | 2021-06-16 | X-ray | UQZ | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.63% | 27.09% | 16.50% | ~{N}-[(1~{S})-2-(1,3-benzodioxol-5-ylmethylamino)-1-(3-hydroxyphenyl)-2-oxidanylidene-ethyl]-~{N}-propyl-prop-2-enamide | ~{N}-[(1~{S})-2-(1,3-benzodioxol-5-ylmethylamino)-1-(3-hydroxyphenyl)-2-oxidanylidene-ethyl]-~{N}-propyl-prop-2-enamide | Aimless | 99.80% | 9.3 | SLS (X06SA) | REFMAC | 2021-03-08 | 34097796 | - | - | - | - | N/A | - | - | 3C-like proteinase | 33.8 | - | No | - | 19.1 | 78.5 | 46.0 | - | ||||||
| 7ntv | 2.06 Å | 2021-06-16 | X-ray | US8 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.49% | 22.75% | 15.80% | 2-acetamido-N-cyclopropyl-5-phenyl-thiophene-3-carboxamide | 2-acetamido-N-cyclopropyl-5-phenyl-thiophene-3-carboxamide | XDS | 99.82% | 11.0 | SLS (X06SA) | REFMAC | 2021-03-10 | 34097796 | - | - | - | - | N/A | - | - | 3C-like proteinase | 58.7 | - | No | - | 57.6 | 59.8 | 74.4 | - | ||||||
| 7nuk | 2.19 Å | 2021-06-16 | X-ray | USH | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.55% | 26.08% | 25.40% | 2-[2-chloranylethanoyl(propyl)amino]-~{N}-(2-methoxyphenyl)ethanamide | 2-[2-chloranylethanoyl(propyl)amino]-~{N}-(2-methoxyphenyl)ethanamide | XDS | 99.30% | 9.6 | SLS (X06SA) | REFMAC | 2021-03-12 | 34097796 | - | - | - | - | N/A | - | - | 3C-like proteinase | 38.5 | - | No | - | 26.1 | 60.1 | 66.8 | - | ||||||
| 7cut | 1.82 Å | 2021-06-23 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.23% | 22.32% | N/A | Z-VAD(OMe)-FMK | - | XDS | 97.00% | 14.0 | SSRF (BL17U1) | PHENIX | 2020-08-24 | 34075025 | - | - | - | - | N/A | - | - | Z-VAD(OMe)-FMK; 3C protein | 48.8 | - | No | - | 61.9 | 55.8 | 55.1 | - | ||||||
| 7cuu | 1.68 Å | 2021-06-23 | X-ray | ALD | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.46% | 19.63% | N/A | N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | XDS | 95.80% | 6.1 | SSRF (BL19U1) | PHENIX | 2020-08-24 | 34075025 | - | - | - | - | N/A | - | - | 3C protein | 68.5 | - | No | - | 84.1 | 60.2 | 68.2 | - | ||||||
| 7cwt | 3.70 Å | 2021-06-23 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2020-08-31 | 33262454 | - | - | - | - | N/A | - | - | Light chain Fab of FC05; Spike glycoprotein; Light chain Fab of HB27; Heavy chain Fab of HB27; Heavy chain Fab of FC05 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7e18 | 1.65 Å | 2021-06-23 | X-ray | HUR | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.40% | 19.92% | 5.20% | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | XDS | 99.30% | 20.8 | Photon Factory (BL-17A) | PHENIX | 2021-02-01 | 34313428 | - | - | - | - | N/A | - | - | Replicase polyprotein 1ab | 68.6 | - | No | - | 82.2 | 37.2 | 93.3 | - | ||||||
| 7e19 | 2.15 Å | 2021-06-23 | X-ray | HUO | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.13% | 23.46% | 23.10% | (phenylmethyl) N-[(2S)-1-[[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]amino]-3-methyl-1-oxidanylidene-butan-2-yl]carbamate | (phenylmethyl) N-[(2S)-1-[[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]amino]-3-methyl-1-oxidanylidene-butan-2-yl]carbamate | XDS | 100.00% | 12.63 | Photon Factory (BL-17A) | PHENIX | 2021-02-01 | 34313428 | - | - | - | - | N/A | - | - | 3C-like proteinase | 69.6 | - | No | - | 50.4 | 73.6 | 90.6 | - | ||||||
| 7e5y | 3.59 Å | 2021-06-23 | X-ray | - | SARS-CoV-2 | Spike | No functional ligands | 23.17% | 27.65% | 15.00% | - | xia2 | 99.90% | 10.9 | SSRF (BL17U1) | PHENIX | 2021-02-21 | 34315862 | - | - | - | - | N/A | - | - | Spike protein S1; 2B11 Fab Heavy chain; 2B11 Fab Light chain | 25.3 | - | No | - | 15.9 | 93.3 | 17.0 | - | |||||||
| 7eaz | 3.50 Å | 2021-06-23 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-03-08 | 34563540 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7eb0 | 3.60 Å | 2021-06-23 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-03-08 | 34563540 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7eb3 | 3.60 Å | 2021-06-23 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-03-08 | 34563540 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7eb4 | 3.50 Å | 2021-06-23 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-03-08 | 34563540 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7eb5 | 3.40 Å | 2021-06-23 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-03-08 | 34563540 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7ej4 | 3.60 Å | 2021-06-23 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-04-01 | - | - | - | - | - | N/A | - | - | RBD-chAb-25, Heavy chain; Spike glycoprotein; RBD-chAb-25, Light chain | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7ej5 | 3.50 Å | 2021-06-23 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-04-01 | - | - | - | - | - | N/A | - | - | RBD-chAb45, Light chain; Spike glycoprotein; RBD-chAb45, Heavy chain | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7mgr | 1.94 Å | 2021-06-23 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Protein-protein complex | 19.33% | 22.86% | 8.50% | ALA-VAL-LYS-LEU-GLN-ASN-ASN-GLU | - | XDS | 97.20% | 8.13 | APS (24-ID-C) | PHENIX | 2021-04-13 | 34437808 | - | - | - | - | N/A | - | - | ALA-VAL-LYS-LEU-GLN-ASN-ASN-GLU; 3C-like proteinase | 62.8 | - | No | - | 56.5 | 79.0 | 64.8 | - | ||||||
| 7mgs | 1.84 Å | 2021-06-23 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 19.34% | 22.73% | 6.30% | SER-ALA-VAL-LEU-GLN-SER-GLY-PHE | - | XDS | 92.50% | 9.8 | APS (24-ID-C) | PHENIX | 2021-04-13 | 34437808 | - | - | - | - | N/A | - | - | 3C-like proteinase; SER-ALA-VAL-LEU-GLN-SER-GLY-PHE | 62.8 | - | No | - | 57.8 | 60.8 | 81.6 | - | ||||||
| 7n89 | 2.00 Å | 2021-06-23 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.06% | 21.80% | 10.30% | ACE-SER-ALA-VAL-LEU-GLN-SER-GLY-PHE-NH2 | - | CrysalisPro | 93.60% | 8.2 | ROTATING ANODE () | PHENIX | 2021-06-14 | 34804549 | - | - | - | - | N/A | - | - | 3C-like proteinase; ACE-SER-ALA-VAL-LEU-GLN-SER-GLY-PHE-NH2 | 72.1 | - | No | - | 66.9 | 67.4 | 86.3 | - | ||||||
| 7n8c | 2.20 Å | 2021-06-23 | X-ray/N-diffr. | YD1, DOD | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.70% | 19.20% | 10.70% | 6-[4-(3,4-dichlorophenyl)piperazin-1-yl]carbonyl-1~{H}-pyrimidine-2,4-dione | 6-[4-(3,4-dichlorophenyl)piperazin-1-yl]carbonyl-1~{H}-pyrimidine-2,4-dione; water | Mantid; CrysalisPro | 96.30% | 15.6 | ROTATING ANODE, SPALLATION SOURCE () | nCNS | 2021-06-14 | 34705466 | - | - | - | - | N/A | - | - | 3C-like proteinase | N/A | - | No | - | N/A | N/A | N/A | - | ||||||
| 7nh7 | 2.20 Å | 2021-06-23 | X-ray | SFG | NSP10/NSP16 | Functional ligand | 16.42% | 21.12% | 13.05% | SINEFUNGIN | SINEFUNGIN | XDS | 99.74% | 11.66 | ROTATING ANODE () | PHENIX; PHENIX | 2021-02-10 | - | ZN | - | - | - | N/A | - | - | Replicase polyprotein 1a; Replicase polyprotein 1ab | 57.4 | - | No | - | - | 73.0 | 69.4 | 46.9 | - | ||||||
| 7dk1 | 1.90 Å | 2021-06-30 | X-ray | BTB | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 18.96% | 20.94% | 8.70% | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL | 100.00% | 2.69 | ELETTRA (11.2C) | BUSTER | 2020-11-22 | 34514483 | ZN | - | - | - | N/A | - | - | 3C-like proteinase | 81.1 | - | No | - | 74.6 | 81.0 | 91.0 | - | |||||||
| 7l4z | 3.96 Å | 2021-06-30 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 25.66% | 28.16% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; ACE-DTY-LYS-ALA-GLY-VAL-VAL-TYR-GLY-TYR-ASN-ALA-TRP-ILE-ARG-CYS-NH2 | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.60% | 7.0 | Australian Synchrotron (MX2) | PHENIX | 2020-12-21 | 34230894 | - | - | - | - | N/A | - | - | ACE-DTY-LYS-ALA-GLY-VAL-VAL-TYR-GLY-TYR-ASN-ALA-TRP-ILE-ARG-CYS-NH2; Spike protein S1 - Receptor-binding domain (RBD) | 19.0 | - | No | - | 13.4 | 53.4 | 45.3 | - | ||||||
| 7m8m | 1.78 Å | 2021-06-30 | X-ray | YSG | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.56% | 23.43% | N/A | 5-[3-(3-chloro-5-propoxyphenyl)-2-oxo-2H-[1,3'-bipyridin]-5-yl]pyrimidine-2,4(1H,3H)-dione | 5-[3-(3-chloro-5-propoxyphenyl)-2-oxo-2H-[1,3'-bipyridin]-5-yl]pyrimidine-2,4(1H,3H)-dione | XDS | 99.60% | 14.6 | APS (24-ID-E) | PHENIX | 2021-03-30 | 34161756 | - | - | - | - | N/A | - | - | 3C-like proteinase | 25.0 | - | No | - | 50.7 | 34.8 | 40.1 | - | ||||||
| 7m8n | 1.96 Å | 2021-06-30 | X-ray | YSP | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.67% | 22.94% | N/A | 5-(3-{3-chloro-5-[(2-methylphenyl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | 5-(3-{3-chloro-5-[(2-methylphenyl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | XDS | 98.30% | 6.24 | NSLS-II (17-ID-1) | PHENIX | 2021-03-30 | 34161756 | - | - | - | - | N/A | - | - | 3C-like proteinase | 27.2 | - | No | - | 55.6 | 39.5 | 34.8 | - | ||||||
| 7m8o | 2.44 Å | 2021-06-30 | X-ray | YSM | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 21.14% | 28.80% | N/A | 5-(3-{3-chloro-5-[(3-fluorophenyl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | 5-(3-{3-chloro-5-[(3-fluorophenyl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | XDS | 99.50% | 11.93 | NSLS-II (17-ID-1) | PHENIX | 2021-03-30 | 34161756 | - | - | - | - | N/A | - | - | 3C-like proteinase | 14.7 | - | No | - | 10.6 | 65.6 | 25.6 | - | ||||||
| 7m8p | 2.23 Å | 2021-06-30 | X-ray | YSJ | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.48% | 23.14% | N/A | 5-(3-{3-chloro-5-[(2-chlorophenyl)methoxy]-4-fluorophenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | 5-(3-{3-chloro-5-[(2-chlorophenyl)methoxy]-4-fluorophenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | XDS | 97.90% | 10.52 | APS (24-ID-E) | PHENIX | 2021-03-30 | 34161756 | - | - | - | - | N/A | - | - | 3C-like proteinase | 55.0 | - | No | - | 53.6 | 86.0 | 45.0 | - | ||||||
| 7m8x | 1.74 Å | 2021-06-30 | X-ray | YTJ | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 22.93% | 27.34% | N/A | 2-{3-[3-chloro-5-(2-methoxyethoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}benzonitrile | 2-{3-[3-chloro-5-(2-methoxyethoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}benzonitrile | XDS | 98.00% | 9.88 | APS (24-ID-E) | PHENIX | 2021-03-30 | 34161756 | - | - | - | - | N/A | - | - | 3C-like proteinase | 11.5 | - | No | - | 17.6 | 26.0 | 49.6 | - | ||||||
| 7m8y | 1.75 Å | 2021-06-30 | X-ray | YTM | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.50% | 22.78% | N/A | 5-{3-[3-chloro-5-(2-phenylethoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}pyrimidine-2,4(1H,3H)-dione | 5-{3-[3-chloro-5-(2-phenylethoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}pyrimidine-2,4(1H,3H)-dione | XDS | 96.30% | 8.07 | APS (24-ID-E) | PHENIX | 2021-03-30 | 34161756 | - | - | - | - | N/A | - | - | 3C-like proteinase | 40.7 | - | No | - | 57.4 | 60.1 | 39.8 | - | ||||||
| 7m8z | 1.79 Å | 2021-06-30 | X-ray | YTV | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.77% | 24.03% | N/A | 5-{3-[3-chloro-5-(3-hydroxy-3-methylbutoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}pyrimidine-2,4(1H,3H)-dione | XDS | 97.10% | 7.87 | APS (24-ID-E) | PHENIX | 2021-03-30 | 34161756 | - | - | - | - | N/A | - | - | 3C-like proteinase | 23.2 | - | No | - | 44.5 | 32.2 | 44.9 | - | |||||||
| 7m90 | 2.19 Å | 2021-06-30 | X-ray | YTS | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 21.64% | 27.26% | N/A | 5-(3-{3-chloro-5-[2-(3-oxopiperazin-1-yl)ethoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | 5-(3-{3-chloro-5-[2-(3-oxopiperazin-1-yl)ethoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | XDS | 97.10% | 4.83 | NSLS-II (17-ID-1) | PHENIX | 2021-03-30 | 34161756 | - | - | - | - | N/A | - | - | 3C-like proteinase | 11.0 | - | No | - | 18.1 | 30.3 | 43.2 | - | ||||||
| 7m91 | 1.95 Å | 2021-06-30 | X-ray | YU4 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.81% | 23.28% | N/A | 5-{3-[3-chloro-5-(3,3,3-trifluoropropoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}pyrimidine-2,4(1H,3H)-dione | 5-{3-[3-chloro-5-(3,3,3-trifluoropropoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}pyrimidine-2,4(1H,3H)-dione | XDS | 96.90% | 10.85 | APS (24-ID-E) | PHENIX | 2021-03-30 | 34161756 | - | - | - | - | N/A | - | - | 3C-like proteinase | 28.4 | - | No | - | 52.3 | 26.3 | 53.9 | - | ||||||
| 7aeg | 1.70 Å | 2021-07-07 | X-ray | F2G | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.78% | 20.04% | 22.40% | 3-(2-{2-[(CYCLOHEXYLMETHOXY-HYDRO-METHYL)-AMINO]-1-HYDROXY-3-METHYL-BUTYLAMINO}-1-HYDROXY-PROPYLAMINO)-PENTANE-1,1,4-TRIOL | 3-(2-{2-[(CYCLOHEXYLMETHOXY-HYDRO-METHYL)-AMINO]-1-HYDROXY-3-METHYL-BUTYLAMINO}-1-HYDROXY-PROPYLAMINO)-PENTANE-1,1,4-TRIOL | xia2; DIALS | 99.90% | 7.6 | Diamond (I24) | REFMAC | 2020-09-17 | 34168183 | - | - | - | - | N/A | - | - | Replicase polyprotein 1ab | 78.2 | - | No | - | 80.2 | 68.7 | 88.0 | - | ||||||
| 7aeh | 1.30 Å | 2021-07-07 | X-ray | R8H | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 14.04% | 17.56% | 19.90% | (2~{R})-5-oxidanylidene-~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(pyridin-2-ylmethylamino)butan-2-yl]-1-(phenylmethyl)pyrrolidine-2-carboxamide | (2~{R})-5-oxidanylidene-~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(pyridin-2-ylmethylamino)butan-2-yl]-1-(phenylmethyl)pyrrolidine-2-carboxamide | xia2; DIALS | 97.20% | 10.7 | Diamond (I04-1) | REFMAC | 2020-09-17 | 34168183 | - | - | - | - | N/A | - | - | 2'-O-methyltransferase | 74.4 | - | No | - | 94.2 | 52.2 | 80.0 | - | ||||||
| 7ein | 1.70 Å | 2021-07-07 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 23.50% | 26.02% | 6.30% | leupeptin | - | HKL-2000 | 100.00% | 29.9 | SSRF (BL17U) | PHENIX; PHENIX | 2021-03-31 | 34579576 | - | - | - | - | N/A | - | - | 3C-like proteinase; leupeptin | 29.4 | - | No | - | 26.4 | 38.3 | 70.1 | - | ||||||
| 7k0e | 1.90 Å | 2021-07-07 | X-ray | K36, PG4 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.09% | 23.02% | 7.40% | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; TETRAETHYLENE GLYCOL | XDS | 98.60% | 8.8 | APS (17-ID) | PHENIX | 2020-09-04 | 34210738 | - | - | - | - | N/A | - | - | 3C-like proteinase - Full Length | 75.6 | - | No | - | 54.8 | 88.4 | 86.2 | - | ||||||
| 7k0g | 1.85 Å | 2021-07-07 | X-ray | B1S, K36 | SARS-CoV | NSP5 (3CLpro) | Functional ligand | 19.17% | 23.95% | 7.60% | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | XDS | 100.00% | 13.7 | APS (17-ID) | PHENIX | 2020-09-04 | 34210738 | - | - | - | - | N/A | - | - | Replicase polyprotein 1a | 54.5 | - | No | - | 45.4 | 59.0 | 79.2 | - | ||||||
| 7k0h | 1.70 Å | 2021-07-07 | X-ray | VR4, PG4 | SARS-CoV | NSP5 (3CLpro) | Functional ligand | 17.43% | 21.00% | 6.70% | N-{(2S,3R)-4-(benzylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-N~2~-[(benzyloxy)carbonyl]-L-leucinamide | N-{(2S,3R)-4-(benzylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-N~2~-[(benzyloxy)carbonyl]-L-leucinamide; TETRAETHYLENE GLYCOL | XDS | 99.90% | 10.5 | APS (17-ID) | PHENIX | 2020-09-04 | 34210738 | - | - | - | - | N/A | - | - | Replicase polyprotein 1a - Full Length | 78.1 | - | No | - | 74.0 | 80.8 | 81.9 | - | ||||||
| 7lco | 1.90 Å | 2021-07-07 | X-ray | XTJ | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.81% | 24.91% | N/A | (3-fluorophenyl)methyl [(2S)-3-cyclopropyl-1-oxo-1-({(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)propan-2-yl]carbamate | (3-fluorophenyl)methyl [(2S)-3-cyclopropyl-1-oxo-1-({(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)propan-2-yl]carbamate | XDS | 95.76% | 22.66 | CLSI (08B1-1) | PHENIX | 2021-01-11 | 34118724 | - | - | - | - | N/A | - | - | 3C-like proteinase | 26.8 | - | No | - | 36.0 | 40.0 | 53.3 | - | ||||||
| 7lcs | 1.85 Å | 2021-07-07 | X-ray | XTP | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.38% | 25.50% | N/A | benzyl [(2S)-3-cyclopropyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamate | benzyl [(2S)-3-cyclopropyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamate | XDS | 96.41% | 12.16 | SSRL (BL12-2) | PHENIX | 2021-01-11 | 34118724 | - | - | - | - | N/A | - | - | 3C-like proteinase | 43.8 | - | No | - | 30.7 | 78.7 | 53.9 | - | ||||||
| 7lct | 1.93 Å | 2021-07-07 | X-ray | XU4 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.00% | 23.32% | N/A | N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-{[(1S)-1-phenylethoxy]carbonyl}-L-leucinamide | N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-{[(1S)-1-phenylethoxy]carbonyl}-L-leucinamide | XDS | 99.34% | 9.34 | SSRL (BL12-2) | PHENIX | 2021-01-11 | 34118724 | - | - | - | - | N/A | - | - | 3C-like proteinase | 44.9 | - | No | - | 51.8 | 45.7 | 67.8 | - | ||||||
| 7ldl | 2.00 Å | 2021-07-07 | X-ray | XV4 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.68% | 26.11% | N/A | N-[(2S)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | N-[(2S)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | XDS | 94.04% | 7.99 | CLSI (08B1-1) | PHENIX | 2021-01-13 | 34118724 | - | - | - | - | N/A | - | - | 3C-like proteinase | 11.3 | - | No | - | 25.7 | 12.7 | 54.0 | - | ||||||
| 7n1q | 3.44 Å | 2021-07-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | 42.23% | 42.48% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-05-28 | 34168070 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7n1t | 3.44 Å | 2021-07-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | 42.23% | 42.48% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-05-28 | 34168070 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7n1u | 3.14 Å | 2021-07-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-05-28 | 34168070 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7n1v | 3.50 Å | 2021-07-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | 43.51% | 43.66% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-05-28 | 34168070 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7n1w | 3.33 Å | 2021-07-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-05-28 | 34168070 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7n1x | 4.00 Å | 2021-07-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-05-28 | 34168070 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7n1y | 4.30 Å | 2021-07-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-05-28 | 34168070 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7n3c | 1.82 Å | 2021-07-07 | X-ray | IOD, PO4 | SARS-CoV-2 | Nucleocapsid | Pathogen-host interaction | 17.94% | 20.97% | 17.20% | PHOSPHATE ION; IODIDE ION | IODIDE ION; PHOSPHATE ION | HKL-3000 | 99.50% | 14.1 | APS (19-ID) | PHENIX | 2021-05-31 | - | - | - | - | - | N/A | - | - | S24-202 Fab heavy chain; S24-202 Fab light chain; Nucleoprotein - CoV N NTD domain, residues 47-173 | 58.4 | - | No | - | - | 74.4 | 46.3 | 70.6 | - | |||||
| 7n3d | 1.53 Å | 2021-07-07 | X-ray | - | SARS-CoV-2 | Nucleocapsid | Pathogen-host interaction | 15.72% | 19.02% | 11.40% | - | HKL-3000 | 95.90% | 22.0 | APS (19-ID) | PHENIX | 2021-05-31 | - | - | - | - | - | N/A | - | - | S24-1564 Fab light chain; Nucleoprotein - CoV N NTD domain, residues 47-173; S24-1564 Fab heavy chain | 76.4 | - | No | - | - | 87.6 | 65.6 | 78.4 | - | ||||||
| 7nw2 | 2.10 Å | 2021-07-07 | X-ray | USZ | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.23% | 22.36% | 8.40% | ~{N}-(4-~{tert}-butylphenyl)-~{N}-[(1~{R})-2-[2-(3-fluorophenyl)ethylamino]-2-oxidanylidene-1-pyridin-3-yl-ethyl]propanamide | ~{N}-(4-~{tert}-butylphenyl)-~{N}-[(1~{R})-2-[2-(3-fluorophenyl)ethylamino]-2-oxidanylidene-1-pyridin-3-yl-ethyl]propanamide | XDS | 95.10% | 14.7 | Diamond (I04-1) | BUSTER; REFMAC | 2021-03-16 | 34174194 | - | - | - | - | N/A | - | - | 3C-like proteinase | 63.0 | - | No | - | 61.6 | 68.5 | 70.5 | - | ||||||
| 7or9 | 2.34 Å | 2021-07-07 | X-ray | PG4 | SARS-CoV-2 | Spike | Pathogen-host interaction | 19.35% | 22.96% | 10.00% | TETRAETHYLENE GLYCOL | xia2 | 86.20% | 16.8 | Diamond (I03) | PHENIX | 2021-06-04 | 34242578 | - | - | - | - | N/A | - | - | COVOX-278 Fab light chain; COVOX-222 Fab heavy chain; Spike protein S1; COVOX-222 Fab light chain; COVOX-278 Fab heavy chain | 63.9 | - | No | - | 55.5 | 68.5 | 78.6 | - | |||||||
| 7ora | 2.60 Å | 2021-07-07 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 20.40% | 24.48% | 24.60% | - | xia2 | 90.50% | 7.3 | Diamond (I03) | PHENIX | 2021-06-04 | 34242578 | - | - | - | - | N/A | - | - | COVOX-45 Fab light chain; Spike protein S1; COVOX-45 Fab heavy chain; COVOX-253 Fab heavy chain; COVOX-253 Fab light chain | 30.1 | - | No | - | 40.4 | 18.7 | 77.0 | - | |||||||
| 7orb | 2.50 Å | 2021-07-07 | X-ray | TRS, BTB, PO4 | SARS-CoV-2 | Spike | Pathogen-host interaction | 21.28% | 25.13% | 30.00% | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL; PHOSPHATE ION; 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL; 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL; PHOSPHATE ION | xia2 | 99.80% | 5.6 | Diamond (I03) | PHENIX | 2021-06-04 | 34242578 | - | - | - | - | N/A | - | - | Spike protein S1; COVOX-253 Fab heavy chain; COVOX-253 Fab light chain; COVOX-75 Fab heavy chain; COVOX-75 Fab light chain | 41.0 | - | No | - | 34.0 | 57.8 | 66.0 | - | ||||||
| 7r98 | 2.51 Å | 2021-07-07 | X-ray | - | SARS-CoV-2 | Nucleocapsid | Pathogen-host interaction | 22.64% | 27.32% | 21.40% | - | XDS | 100.00% | 9.7 | APS (24-ID-C) | PHENIX | 2021-06-28 | 34381460 | - | - | - | - | N/A | - | - | Nucleoprotein - RNA-binding domain; Nanobody B6 | 10.1 | - | No | - | 17.7 | 22.2 | 48.9 | - | |||||||
| 7lrs | 3.89 Å | 2021-07-14 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-02-17 | 34210892 | - | - | - | - | N/A | - | - | antibody A23-58.1 light chain; antibody A23-58.1 heavy chain; Spike glycoprotein - receptor binding domain (UNP residues 332-527) | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lrt | 3.54 Å | 2021-07-14 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-02-17 | 34210892 | - | - | - | - | N/A | - | - | SARS-CoV-2 spike glycoprotein; antibody A23-58.1 light chain - Fab; antibody A23-58.1 heavy chain - Fab | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7n8h | 2.30 Å | 2021-07-14 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-06-14 | 34210893 | - | - | - | - | N/A | - | - | S2M11 Fab Heavy Chain variable region; S2L20 Fab Heavy Chain variable region; Spike glycoprotein; S2M11 Fab Light Chain variable region; S2L20 Fab Light Chain variable region | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7n8i | 3.00 Å | 2021-07-14 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-06-14 | 34210893 | - | - | - | - | N/A | - | - | Spike glycoprotein; S2L20 Fab Light Chain Variable Region; S2L20 Fab Heavy Chain Variable Region | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7rb0 | 2.98 Å | 2021-07-14 | Cryo-EM | - | SARS-CoV-2 | NSP15 | No functional ligands | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-07-05 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7rb2 | 3.27 Å | 2021-07-14 | Cryo-EM | - | SARS-CoV-2 | NSP15 | No functional ligands | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-07-05 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7bwq | 2.95 Å | 2021-07-21 | X-ray | - | SARS-CoV-2 | NSP9 | No functional ligands | 21.28% | 29.89% | 22.10% | - | HKL-2000 | 100.00% | 6.4 | SSRF (BL18U1) | PHENIX | 2020-04-15 | 35006420 | - | - | - | - | N/A | - | - | Nsp9 | 4.6 | - | No | - | 7.5 | 46.1 | 14.4 | - | |||||||
| 7egq | 3.35 Å | 2021-07-21 | Cryo-EM | - | SARS-CoV-2 | NSP7/NSP8/NSP9/NSP10/NSP12/NSP13/NSP14 | No functional ligands | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-03-25 | 34143953 | MG; ZN | - | - | - | N/A | - | - | Template RNA; primer RNA; Proofreading exoribonuclease - UNP residues 5926-6452; Non-structural protein 10; Non-structural protein 9 - UNP residues 4141-4253; Helicase - UNP residues 5325-5925; Non-structural protein 7; Non-structural protein 8 - UNP residues 3943-4140; RNA-directed RNA polymerase - UNP residues 4393-5324 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7fay | 2.10 Å | 2021-07-21 | X-ray | 2XI | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.29% | 23.36% | 7.60% | (2~{R})-~{N}-[(1~{R})-2-(~{tert}-butylamino)-2-oxidanylidene-1-pyridin-3-yl-ethyl]-~{N}-(4-~{tert}-butylphenyl)-2-oxidanyl-propanamide | (2~{R})-~{N}-[(1~{R})-2-(~{tert}-butylamino)-2-oxidanylidene-1-pyridin-3-yl-ethyl]-~{N}-(4-~{tert}-butylphenyl)-2-oxidanyl-propanamide | XDS | 99.60% | 11.9 | SSRF (BL19U1) | BUSTER | 2021-07-08 | 35477751 | - | - | - | - | N/A | - | - | 3C-like proteinase | 45.9 | - | No | - | 51.5 | 39.4 | 76.2 | - | ||||||
| 7faz | 2.10 Å | 2021-07-21 | X-ray | 2RI, NA | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.03% | 24.28% | 11.80% | SODIUM ION; (2~{R})-~{N}-dibenzofuran-3-yl-~{N}-[(1~{R})-2-[[(1~{S})-1-(4-fluorophenyl)ethyl]amino]-2-oxidanylidene-1-pyridin-3-yl-ethyl]-2-oxidanyl-propanamide | (2~{R})-~{N}-dibenzofuran-3-yl-~{N}-[(1~{R})-2-[[(1~{S})-1-(4-fluorophenyl)ethyl]amino]-2-oxidanylidene-1-pyridin-3-yl-ethyl]-2-oxidanyl-propanamide; SODIUM ION | XDS | 99.80% | 9.4 | SSRF (BL19U1) | PHENIX | 2021-07-08 | 35477751 | - | - | - | - | N/A | - | - | 3C-like proteinase | 42.4 | - | No | - | 42.3 | 45.3 | 72.9 | - | ||||||
| 7kri | 1.58 Å | 2021-07-21 | X-ray | X0Y, MLI | SARS-CoV-2 | NSP9 | Functional ligand | 17.77% | 19.53% | N/A | 1,3-dimethyl-1H-pyrrolo[3,4-d]pyrimidine-2,4(3H,6H)-dione; MALONATE ION | 1,3-dimethyl-1H-pyrrolo[3,4-d]pyrimidine-2,4(3H,6H)-dione; MALONATE ION | HKL-2000 | 100.00% | 13.4 | Australian Synchrotron (MX2) | PHENIX | 2020-11-20 | 34331944 | - | - | - | - | N/A | - | - | Non-structural protein 9 | 51.0 | - | No | - | 84.8 | 18.8 | 73.4 | - | ||||||
| 7mbi | 2.15 Å | 2021-07-21 | X-ray | YWJ, FN7 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.41% | 26.07% | N/A | 4-methoxy-N-[(2S)-4-methyl-1-oxo-1-({(2S)-3-oxo-1-[(3S)-2-oxopiperidin-3-yl]butan-2-yl}amino)pentan-2-yl]-1H-indole-2-carboxamide; 2,4,6-trimethylpyridine-3-carboxylic acid | 4-methoxy-N-[(2S)-4-methyl-1-oxo-1-({(2S)-3-oxo-1-[(3S)-2-oxopiperidin-3-yl]butan-2-yl}amino)pentan-2-yl]-1H-indole-2-carboxamide; 2,4,6-trimethylpyridine-3-carboxylic acid | XDS | 99.69% | 13.73 | SSRL (BL12-1) | PHENIX | 2021-03-31 | 34242027 | - | - | - | - | N/A | - | - | 3C-like proteinase | 19.5 | - | No | - | 26.1 | 46.4 | 40.7 | - | ||||||
| 7n44 | 1.94 Å | 2021-07-21 | X-ray | 06I | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.64% | 21.85% | N/A | 5-(3-{3-chloro-5-[(5-methyl-1,3-thiazol-4-yl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | 5-(3-{3-chloro-5-[(5-methyl-1,3-thiazol-4-yl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | XDS | 96.73% | 12.79 | APS (24-ID-E) | PHENIX | 2021-06-03 | 34408808 | - | - | - | - | N/A | - | - | 3C-like proteinase | 69.3 | - | No | - | 66.5 | 83.2 | 64.4 | - | ||||||
| 7p35 | 2.26 Å | 2021-07-21 | X-ray | AG7 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.00% | 25.81% | N/A | 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | XDS | 84.40% | 9.6 | ALBA (XALOC) | REFMAC | 2021-07-07 | 36336176 | - | - | - | - | N/A | - | - | 3C-like proteinase | 37.1 | - | No | - | 28.1 | 73.6 | 48.6 | - | |||||||
| 7r6w | 1.83 Å | 2021-07-21 | X-ray | POL | SARS-CoV-2 | Spike | Pathogen-host interaction | 21.13% | 23.16% | 8.50% | N-PROPANOL | N-PROPANOL | XDS | 99.60% | 16.2 | SSRL (BL9-2) | REFMAC | 2021-06-23 | 34261126 | - | - | - | - | N/A | - | - | Heavy Chain of Fab domain of monoclonal antibody S309; Spike protein S1 - receptor-binding domain (UNP reisdues 328-531); Heavy Chain of Fab domain of monoclonal antibody S2X35; Light Chain of Fab domain of monoclonal antibody S2X35; Light Chain of Fab domain of monoclonal antibody S309 | 69.4 | - | No | - | 53.5 | 66.4 | 94.4 | - | ||||||
| 7r6x | 2.95 Å | 2021-07-21 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 23.23% | 26.24% | 29.50% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 100.00% | 13.3 | ALS (4.2.2) | REFMAC | 2021-06-23 | 34261126 | - | - | - | - | N/A | - | - | Spike protein S1 - receptor-binding domain (UNP residues 328-531); Monoclonal antibody S309 Fab heavy chain; Monoclonal antibody S309 Fab light chain; Monoclonal antibody S2E12 Fab heavy chain; Monoclonal antibody S2E12 Fab light chain; Monoclonal antibody S304 Fab light chain; Monoclonal antibody S304 Fab heavy chain | 6.9 | - | No | - | 24.9 | 15.1 | 37.8 | - | ||||||
| 7r7n | 3.95 Å | 2021-07-21 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | 2021-06-25 | 34261126 | - | - | - | - | N/A | - | - | Spike glycoprotein; S2D106 FAB light chain; S2D106 FAB heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7m42 | 3.30 Å | 2021-07-28 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | PHENIX; phenix.real_space_refine | 2021-03-19 | 34161776 | - | - | - | - | N/A | - | - | REGN10989 antibody Fab fragment heavy chain; REGN10985 antibody Fab fragment heavy chain; REGN10985 antibody Fab fragment light chain; Spike protein S1 - receptor binding domain (UNP residues 319-541); REGN10989 antibody Fab fragment light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7mlz | 3.71 Å | 2021-07-28 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-04-29 | 34210892 | - | - | - | - | N/A | - | - | B1-182.1 Fab heavy chain; Spike protein S1 - Receptor binding domain, UNP residues 331-527; B1-182.1 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7mm0 | 3.15 Å | 2021-07-28 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-04-29 | 34210892 | - | - | - | - | N/A | - | - | Spike glycoprotein; B1-182.1 Fab light chain; B1-182.1 Fab heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7n0b | 3.90 Å | 2021-07-28 | Cryo-EM | CA | SARS-CoV-2 | NSP10/NSP14 | Protein-protein complex | N/A | N/A | N/A | RNA (5'-R(*AP*GP*AP*AP*GP*CP*UP*AP*UP*UP*AP*AP*AP*AP*UP*CP*AP*CP*C)-3'); CALCIUM ION; RNA (5'-R(*AP*UP*GP*UP*GP*AP*UP*UP*UP*UP*AP*AP*UP*AP*GP*CP*UP*UP*CP*U)-3') | CALCIUM ION | - | - | () | PHENIX | 2021-05-25 | 34315827 | CA; ZN | - | - | - | N/A | - | - | Non-structural protein 10 - UNP residues 4254-4392; RNA (5'-R(*AP*GP*AP*AP*GP*CP*UP*AP*UP*UP*AP*AP*AP*AP*UP*CP*AP*CP*C)-3'); RNA (5'-R(*AP*UP*GP*UP*GP*AP*UP*UP*UP*UP*AP*AP*UP*AP*GP*CP*UP*UP*CP*U)-3'); Proofreading exoribonuclease - UNP residues 5926-6452 | N/A | - | No | - | N/A | N/A | N/A | RNA (5'-R(*AP*GP*AP*AP*GP*CP*UP*AP*UP*UP*AP*AP*AP*AP*UP*CP*AP*CP*C)-3'); RNA (5'-R(*AP*UP*GP*UP*GP*AP*UP*UP*UP*UP*AP*AP*UP*AP*GP*CP*UP*UP*CP*U)-3') | - | ||||||
| 7n0c | 3.40 Å | 2021-07-28 | Cryo-EM | - | SARS-CoV-2 | NSP10/NSP14 | Protein-protein complex | N/A | N/A | N/A | RNA (25-MER); RNA (24-MER) | - | - | - | () | PHENIX | 2021-05-25 | 34315827 | ZN; MG | - | - | - | N/A | - | - | Non-structural protein 10 - UNP residues 4254-4392; RNA (24-MER); RNA (25-MER); Proofreading exoribonuclease - UNP residues 5926-6452 | N/A | - | No | - | N/A | N/A | N/A | RNA (25-MER); RNA (24-MER) | - | ||||||
| 7n0d | 2.50 Å | 2021-07-28 | Cryo-EM | 1N7 | SARS-CoV-2 | NSP10/NSP14 | Protein-protein complex | N/A | N/A | N/A | RNA (5'-R(*GP*GP*GP*GP*AP*UP*GP*UP*GP*AP*UP*UP*UP*UP*AP*AP*UP*AP*G)-3'); CHAPSO; RNA (5'-R(*CP*CP*CP*CP*C)-3'); RNA (5'-R(*CP*UP*AP*UP*UP*AP*AP*AP*AP*UP*CP*AP*CP*C)-3') | CHAPSO | - | - | () | PHENIX | 2021-05-25 | 34315827 | ZN; MG | - | - | - | N/A | - | - | RNA (5'-R(*CP*CP*CP*CP*C)-3'); Non-structural protein 10 - UNP residues 4254-4392; Proofreading exoribonuclease - UNP residues 5926-6452; RNA (5'-R(*GP*GP*GP*GP*AP*UP*GP*UP*GP*AP*UP*UP*UP*UP*AP*AP*UP*AP*G)-3'); RNA (5'-R(*CP*UP*AP*UP*UP*AP*AP*AP*AP*UP*CP*AP*CP*C)-3') | N/A | - | No | - | N/A | N/A | N/A | RNA (5'-R(*GP*GP*GP*GP*AP*UP*GP*UP*GP*AP*UP*UP*UP*UP*AP*AP*UP*AP*G)-3'); RNA (5'-R(*CP*CP*CP*CP*C)-3'); RNA (5'-R(*CP*UP*AP*UP*UP*AP*AP*AP*AP*UP*CP*AP*CP*C)-3') | - | ||||||
| 7f62 | 3.60 Å | 2021-08-04 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-06-24 | 34673836 | - | - | - | - | N/A | - | - | RBD-chAb-25, Light chain; RBD-chAb-25, Heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7f63 | 3.90 Å | 2021-08-04 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-06-24 | 34673836 | - | - | - | - | N/A | - | - | RBD-chAb45, Heavy chain; Spike glycoprotein; RBD-chAb45, Light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7km5 | 3.19 Å | 2021-08-04 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 24.68% | 29.41% | 14.50% | - | XDS | 98.30% | 8.2 | APS (24-ID-E) | PHENIX | 2020-11-02 | 34338634 | - | - | - | - | N/A | - | - | Spike protein S1 - receptor-binding domain; nanobody | 20.7 | - | No | - | 8.7 | 56.6 | 50.7 | - | |||||||
| 7n3i | 2.03 Å | 2021-08-04 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 17.90% | 20.82% | 9.70% | 2-acetamido-2-deoxy-beta-D-glucopyranose | MOSFLM | 96.00% | 6.8 | SSRL (BL12-2) | PHENIX | 2021-06-01 | 34331873 | - | - | - | - | N/A | - | - | C098 Fab heavy chain; Spike protein S1 - Receptor Binding Domain; C098 Fab light chain | 52.1 | - | No | - | 75.5 | 27.1 | 76.5 | - | |||||||
| 7n62 | 4.00 Å | 2021-08-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-06-07 | 33758863 | - | - | - | - | N/A | - | - | C12C9 Fab light chain; C12C9 Fab heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7n64 | 4.20 Å | 2021-08-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-06-07 | 33758863 | - | - | - | - | N/A | - | - | G32R7 Fab heavy chain; Spike glycoprotein; G32R7 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7n9a | 3.50 Å | 2021-08-04 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | 2021-06-17 | - | - | - | - | - | N/A | - | - | Spike protein S1 - Receptor Binding Domain; Nanobody Nb21 | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7n9b | 3.80 Å | 2021-08-04 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | 2021-06-17 | - | - | - | - | - | N/A | - | - | NB21 Nanobody; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7n9c | 3.71 Å | 2021-08-04 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | 2021-06-17 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; Nanobody NB95 | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7n9e | 3.52 Å | 2021-08-04 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | 2021-06-17 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; Nb34 nanobody | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7p77 | 2.98 Å | 2021-08-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-07-19 | 35253970 | - | - | - | - | N/A | - | - | sybody#68; Spike glycoprotein; sybody#15 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7p78 | 3.32 Å | 2021-08-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-07-19 | 35253970 | - | - | - | - | N/A | - | - | sybody#68; Spike glycoprotein; sybody#15 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7p79 | 4.00 Å | 2021-08-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-07-19 | 35253970 | - | - | - | - | N/A | - | - | sybody#15; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7p7a | 4.76 Å | 2021-08-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-07-19 | 35253970 | - | - | - | - | N/A | - | - | sybody#68; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7p7b | 3.13 Å | 2021-08-04 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-07-19 | 35253970 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7r8l | 2.60 Å | 2021-08-04 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 17.97% | 22.86% | 25.70% | beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | - | XDS | 100.00% | 8.9 | SSRL (BL12-2) | PHENIX | 2021-06-26 | 34331873 | - | - | - | - | N/A | - | - | CR3022 Fab light chain; C099 Fab Heavy Chain; C099 Fab Light Chain; Spike protein S1; CR3022 Fab heavy chain | 56.3 | - | No | - | 56.5 | 81.4 | 49.3 | - | ||||||
| 7r8m | 3.40 Å | 2021-08-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-06-26 | 34331873 | - | - | - | - | N/A | - | - | Spike glycoprotein; C032 Fab Heavy Chain; C032 Fab Light Chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7r8n | 3.55 Å | 2021-08-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-06-26 | 34331873 | - | - | - | - | N/A | - | - | C051 Fab Heavy Chain; C051 Fab Light Chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7r8o | 3.50 Å | 2021-08-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-06-26 | 34331873 | - | - | - | - | N/A | - | - | Spike glycoprotein; C548 Fab Light Chain; C548 Fab Heavy Chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7ra8 | 3.10 Å | 2021-08-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-06-30 | 34280951 | - | - | - | - | N/A | - | - | Spike glycoprotein; Light chain Fab S2X259 Fab variable domain; Heavy chain Fab S2X259 Fab variable domain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ral | 3.20 Å | 2021-08-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-07-01 | 34280951 | - | - | - | - | N/A | - | - | S2X259 Fab light chain; Spike glycoprotein; S2X259 Fab heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7lmd | 1.96 Å | 2021-08-11 | X-ray | Y6A | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.77% | 23.35% | 6.00% | 2-(benzotriazol-1-yl)-~{N}-[4-(1~{H}-pyrazol-4-yl)phenyl]-~{N}-(thiophen-3-ylmethyl)ethanamide | 2-(benzotriazol-1-yl)-~{N}-[4-(1~{H}-pyrazol-4-yl)phenyl]-~{N}-(thiophen-3-ylmethyl)ethanamide | HKL-2000 | 98.68% | 4.7 | APS (21-ID-F) | PHENIX | 2021-02-05 | 34347470 | - | - | - | - | N/A | - | - | 3C-like proteinase | 53.9 | - | No | - | 51.6 | 51.9 | 79.1 | - | ||||||
| 7lme | 2.10 Å | 2021-08-11 | X-ray | Y6J | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.01% | 24.05% | 14.00% | ~{N}-[4-[2-(benzotriazol-1-yl)ethanoyl-(thiophen-3-ylmethyl)amino]phenyl]cyclopropanecarboxamide | ~{N}-[4-[2-(benzotriazol-1-yl)ethanoyl-(thiophen-3-ylmethyl)amino]phenyl]cyclopropanecarboxamide | HKL-2000 | 99.16% | 15.1 | APS (21-ID-F) | PHENIX | 2021-02-05 | 34347470 | - | - | - | - | N/A | - | - | 3C-like proteinase | 62.8 | - | No | - | 44.4 | 73.6 | 82.3 | - | ||||||
| 7lmf | 2.20 Å | 2021-08-11 | X-ray | Y6G | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.57% | 25.39% | 13.00% | 2-(benzotriazol-1-yl)-~{N}-[4-(1~{H}-imidazol-4-yl)phenyl]-~{N}-(thiophen-3-ylmethyl)ethanamide | 2-(benzotriazol-1-yl)-~{N}-[4-(1~{H}-imidazol-4-yl)phenyl]-~{N}-(thiophen-3-ylmethyl)ethanamide | HKL-2000 | 99.53% | 14.1 | APS (21-ID-F) | PHENIX | 2021-02-05 | 34347470 | - | - | - | - | N/A | - | - | 3C-like proteinase | 55.3 | - | No | - | 31.9 | 78.7 | 74.7 | - | ||||||
| 7lmg | 1.60 Å | 2021-08-11 | X-ray | Y6G | SARS-CoV | NSP5 (3CLpro) | Functional ligand | 18.45% | 20.78% | 8.48% | 2-(benzotriazol-1-yl)-~{N}-[4-(1~{H}-imidazol-4-yl)phenyl]-~{N}-(thiophen-3-ylmethyl)ethanamide | 2-(benzotriazol-1-yl)-~{N}-[4-(1~{H}-imidazol-4-yl)phenyl]-~{N}-(thiophen-3-ylmethyl)ethanamide | HKL-2000 | 96.98% | 7.38 | APS (21-ID-F) | REFMAC | 2021-02-05 | 34347470 | - | - | - | - | N/A | - | - | 3C-like proteinase | 65.3 | - | No | - | 76.0 | 73.8 | 55.7 | - | ||||||
| 7lmh | 1.85 Å | 2021-08-11 | X-ray | Y6D | SARS-CoV | NSP5 (3CLpro) | Functional ligand | 17.90% | 21.52% | 1.90% | 2-(benzotriazol-1-yl)-~{N}-(4-pyridin-3-ylphenyl)-~{N}-(thiophen-3-ylmethyl)ethanamide | 2-(benzotriazol-1-yl)-~{N}-(4-pyridin-3-ylphenyl)-~{N}-(thiophen-3-ylmethyl)ethanamide | HKL-2000 | 99.42% | 5.77 | APS (21-ID-F) | REFMAC | 2021-02-05 | 34347470 | - | - | - | - | N/A | - | - | 3C-like proteinase | 67.4 | - | No | - | 69.6 | 73.8 | 66.7 | - | ||||||
| 7lmi | 1.71 Å | 2021-08-11 | X-ray | Y6A | SARS-CoV | NSP5 (3CLpro) | Functional ligand | 16.74% | 21.56% | 5.40% | 2-(benzotriazol-1-yl)-~{N}-[4-(1~{H}-pyrazol-4-yl)phenyl]-~{N}-(thiophen-3-ylmethyl)ethanamide | 2-(benzotriazol-1-yl)-~{N}-[4-(1~{H}-pyrazol-4-yl)phenyl]-~{N}-(thiophen-3-ylmethyl)ethanamide | HKL-2000 | 99.75% | 2.59 | APS (21-ID-F) | REFMAC | 2021-02-05 | 34347470 | - | - | - | - | N/A | - | - | 3C-like proteinase | 53.6 | - | No | - | 69.3 | 64.4 | 48.3 | - | ||||||
| 7lmj | 1.69 Å | 2021-08-11 | X-ray | Y67 | SARS-CoV | NSP5 (3CLpro) | Functional ligand | 14.56% | 19.17% | 2.90% | 2-(benzotriazol-1-yl)-~{N}-[(3-chlorophenyl)methyl]-~{N}-[4-(2-oxidanylidene-1~{H}-pyridin-3-yl)phenyl]ethanamide | 2-(benzotriazol-1-yl)-~{N}-[(3-chlorophenyl)methyl]-~{N}-[4-(2-oxidanylidene-1~{H}-pyridin-3-yl)phenyl]ethanamide | HKL-2000 | 99.50% | 17.06 | APS (21-ID-F) | REFMAC | 2021-02-05 | 34347470 | - | - | - | - | N/A | - | - | 3C-like proteinase | 55.3 | - | No | - | 86.9 | 45.7 | 52.7 | - | ||||||
| 7mdw | 3.58 Å | 2021-08-11 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-04-06 | - | - | - | - | - | N/A | - | - | nanobody Nb105; Spike protein S1; nonobody Nb21 | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7me7 | 3.73 Å | 2021-08-11 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-04-06 | - | - | - | - | - | N/A | - | - | Nanobody Nb105; Spike protein S1; Nanobody Nb17 | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7mej | 3.55 Å | 2021-08-11 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-04-06 | - | - | - | - | - | N/A | - | - | Nanobody Nb21; Nanobody Nb36; Spike protein S1 | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7n9t | 3.18 Å | 2021-08-11 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-06-18 | - | - | - | - | - | N/A | - | - | Nanobody Nb17; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7oan | 3.00 Å | 2021-08-11 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 34.54% | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | REFMAC | 2021-04-19 | 34552091 | - | - | - | - | N/A | - | - | Spike glycoprotein; immunoglobulin mu heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7oao | 1.50 Å | 2021-08-11 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 15.21% | 18.60% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 99.40% | 28.1 | Diamond (I03) | REFMAC | 2021-04-19 | 34552091 | - | - | - | - | N/A | - | - | C5 nanobody; Spike protein S1 | 70.9 | - | No | - | 89.7 | 50.2 | 77.7 | - | ||||||
| 7oap | 1.90 Å | 2021-08-11 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 18.02% | 20.40% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 100.00% | 14.4 | Diamond (I03) | REFMAC | 2021-04-19 | 34552091 | - | - | - | - | N/A | - | - | H3 nanobody; Spike protein S1; C1 nanobody | 52.3 | - | No | - | 78.8 | 16.8 | 83.8 | - | ||||||
| 7oaq | 1.55 Å | 2021-08-11 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 15.50% | 17.83% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 100.00% | 16.9 | Diamond (I04) | REFMAC | 2021-04-20 | 34552091 | - | - | - | - | N/A | - | - | Spike protein S1; H3; H3 | 55.1 | - | No | - | 92.7 | 12.4 | 79.7 | - | ||||||
| 7oau | 1.65 Å | 2021-08-11 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 17.54% | 19.92% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 100.00% | 10.0 | Diamond (I24) | REFMAC | 2021-04-20 | 34552091 | - | - | - | - | N/A | - | - | Spike protein S1; C5 | 81.6 | - | No | - | 82.2 | 84.1 | 82.2 | - | ||||||
| 7oay | 2.34 Å | 2021-08-11 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 19.19% | 22.75% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 100.00% | 9.9 | Diamond (I03) | REFMAC | 2021-04-20 | 34552091 | - | - | - | - | N/A | - | - | F2 nanobody; Spike protein S1 | 60.2 | - | No | - | 57.6 | 71.9 | 65.5 | - | ||||||
| 7olz | 1.75 Å | 2021-08-11 | X-ray | DMX | SARS-CoV-2 | Spike | Functional ligand | 17.57% | 20.92% | 9.04% | 3-[BENZYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE | 3-[BENZYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE | XDS | 85.05% | 17.91 | SLS (X10SA) | PHENIX | 2021-05-20 | 34302370 | - | - | - | - | N/A | - | - | Nanobody Re9F06; Nanobody Re5D06; Spike protein S1 | 73.5 | - | No | - | 74.7 | 66.6 | 82.9 | - | ||||||
| 7rn0 | 2.25 Å | 2021-08-11 | X-ray | 5ZB | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 21.38% | 24.10% | 7.10% | (2R)-2-{acetyl[4-(1H-pyrrol-1-yl)phenyl]amino}-N-[(1S)-1-phenylethyl]-2-(pyridin-3-yl)acetamide | (2R)-2-{acetyl[4-(1H-pyrrol-1-yl)phenyl]amino}-N-[(1S)-1-phenylethyl]-2-(pyridin-3-yl)acetamide | iMOSFLM | 92.40% | 7.6 | APS (19-ID) | REFMAC | 2021-07-28 | 34860011 | - | - | - | - | N/A | - | - | 3C-like proteinase | 49.4 | - | No | - | 43.8 | 88.6 | 41.5 | - | ||||||
| 7cot | 2.16 Å | 2021-08-18 | X-ray | - | SARS-CoV-2 | Spike | No functional ligands | 19.67% | 24.66% | 8.80% | - | HKL-2000 | 98.40% | 13.55 | NSRRC (TPS 05A) | PHENIX | 2020-08-05 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | 16.5 | - | No | - | - | 38.6 | 19.1 | 48.5 | - | ||||||
| 7my8 | - | 2021-08-18 | NMR | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2021-05-20 | 34375093 | - | - | - | - | N/A | - | - | Spike protein S2 - residues 816-857 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7ozu | 3.30 Å | 2021-08-18 | Cryo-EM | - | SARS-CoV-2 | NSP7/NSP8/NSP12 | No functional ligands | N/A | N/A | N/A | Product RNA; Template RNA | - | - | - | () | phenix.real_space_refine; PHENIX | 2021-06-28 | 34381216 | ZN | - | - | - | N/A | - | - | Replicase polyprotein 1ab; Non-structural protein 7; Product RNA; Template RNA; Non-structural protein 8 | N/A | - | No | - | N/A | N/A | N/A | Template RNA; Product RNA | - | ||||||
| 7ozv | 3.20 Å | 2021-08-18 | Cryo-EM | - | SARS-CoV-2 | NSP7/NSP8/NSP12 | No functional ligands | N/A | N/A | N/A | Product RNA; Template RNA | - | - | - | () | PHENIX | 2021-06-28 | 34381216 | ZN | - | - | - | N/A | - | - | Replicase polyprotein 1ab; Non-structural protein 8; Non-structural protein 7; Product RNA; Template RNA | N/A | - | No | - | N/A | N/A | N/A | Product RNA; Template RNA | - | ||||||
| 7rqg | 2.17 Å | 2021-08-18 | X-ray | - | SARS-CoV-2 | NSP3(C-term) | No functional ligands | 19.68% | 24.17% | 10.40% | - | HKL-3000 | 90.50% | 15.26 | APS (19-ID) | PHENIX | 2021-08-06 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 - Y3 domain | 18.6 | - | No | - | - | 43.3 | 19.0 | 48.9 | - | ||||||
| 7drv | 3.09 Å | 2021-08-25 | X-ray | NAG | Bat-CoV-RaTG13 | Spike | Pathogen-host interaction | 22.60% | 25.00% | 10.60% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000; HKL-2000 | 99.50% | 14.3 | SSRF (BL19U1) | PHENIX | 2020-12-29 | 34139177 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein - RBD | 34.5 | - | No | - | 35.1 | 59.6 | 50.3 | - | ||||||
| 7fdg | 3.69 Å | 2021-08-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-07-16 | 34580297 | - | - | - | - | N/A | - | - | Spike protein S1; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7fdh | 3.72 Å | 2021-08-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-07-16 | 34580297 | - | - | - | - | N/A | - | - | Spike protein S1; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7fdi | 3.12 Å | 2021-08-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-07-16 | 34580297 | - | - | - | - | N/A | - | - | Spike protein S1; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7fdk | 3.69 Å | 2021-08-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-07-16 | 34580297 | - | - | - | - | N/A | - | - | Spike protein S1; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7m2p | 1.70 Å | 2021-08-25 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.57% | 20.58% | N/A | Inhibitor 18 in bound form | - | iMOSFLM | 99.90% | 10.2 | ROTATING ANODE () | PHENIX | 2021-03-17 | 34288674 | - | - | - | - | N/A | - | - | Inhibitor 18 in bound form; 3C-like proteinase | 37.0 | - | No | - | 77.6 | 13.1 | 59.3 | - | ||||||
| 7oyg | 5.50 Å | 2021-08-25 | Cryo-EM | - | SARS-CoV-2 | NSP7/NSP8/NSP12 | No functional ligands | N/A | N/A | N/A | RNA (5'-R(P*CP*UP*AP*CP*GP*CP*AP*GP*UP*G)-3'); RNA (5'-R(P*UP*GP*CP*AP*CP*UP*GP*CP*GP*UP*AP*G)-3') | - | - | - | () | 2021-06-24 | - | ZN | - | - | - | N/A | - | - | RNA (5'-R(P*UP*GP*CP*AP*CP*UP*GP*CP*GP*UP*AP*G)-3'); SARS-CoV-2 nsp7; SARS-CoV-2 nsp8; SARS-CoV-2 RNA-dependent RNA polymerase (nsp12); RNA (5'-R(P*CP*UP*AP*CP*GP*CP*AP*GP*UP*G)-3') | N/A | - | No | - | - | N/A | N/A | N/A | RNA (5'-R(P*CP*UP*AP*CP*GP*CP*AP*GP*UP*G)-3'); RNA (5'-R(P*UP*GP*CP*AP*CP*UP*GP*CP*GP*UP*AP*G)-3') | - | ||||||
| 7e39 | 3.70 Å | 2021-09-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-02-08 | 34285195 | - | - | - | - | N/A | - | - | Heavy Chain of Ab4; Light Chain of Ab4; Spike protein S1 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7e3b | 4.20 Å | 2021-09-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-02-08 | 34285195 | - | - | - | - | N/A | - | - | Heavy Chain of Ab5; Spike protein S1; Light Chain of Ab5 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7e3c | 4.20 Å | 2021-09-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-02-08 | 34285195 | - | - | - | - | N/A | - | - | Light chain of Ab1; Heavy chain of Ab1; Spike protein S1 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7edf | 3.20 Å | 2021-09-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-03-16 | 34385690 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7edg | 3.20 Å | 2021-09-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-03-16 | 34385690 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7edh | 3.60 Å | 2021-09-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-03-16 | 34385690 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7edi | 3.30 Å | 2021-09-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-03-16 | 34385690 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7edj | 3.30 Å | 2021-09-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-03-16 | 34385690 | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2 (ACE2) ectodomain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7eh5 | 4.00 Å | 2021-09-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-03-28 | 34385690 | - | - | - | - | N/A | - | - | RBD-chAb15, light chain; RBD-chAb45, light chain; RBD-chAb15, heavy chain; Spike glycoprotein; RBD-chAb45, heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7f2b | 2.00 Å | 2021-09-01 | X-ray | PO4 | SARS-CoV-2 | Nucleocapsid | No functional ligands | 15.66% | 21.35% | 4.60% | PHOSPHATE ION | PHOSPHATE ION | CrysalisPro | 100.00% | 11.79 | ROTATING ANODE () | PHENIX | 2021-06-10 | 34665939 | - | - | - | - | N/A | - | - | Nucleoprotein | 74.1 | - | No | - | 71.1 | 91.4 | 63.0 | - | ||||||
| 7l7d | 2.50 Å | 2021-09-01 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 18.46% | 23.10% | 4.60% | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.90% | 19.2 | APS (21-ID-F) | PHENIX | 2020-12-28 | 34548634 | - | - | - | - | N/A | - | - | light chain of human monoclonal antibody Fab AZD8895; heavy chain of human monoclonal antibody Fab AZD8895; Spike protein S1 - receptor binding domain (UNP residues 330-529) | 48.6 | - | No | - | 53.9 | 42.4 | 76.0 | - | |||||||
| 7l7e | 3.00 Å | 2021-09-01 | X-ray | NAG, PO4 | SARS-CoV-2 | Spike | Pathogen-host interaction | 21.54% | 27.29% | 23.10% | PHOSPHATE ION | 2-acetamido-2-deoxy-beta-D-glucopyranose; PHOSPHATE ION | XDS | 99.90% | 6.4 | APS (21-ID-G) | PHENIX | 2020-12-28 | 34548634 | - | - | - | - | N/A | - | - | light chain of human monoclonal antibody Fab AZD8895; Spike protein S1 - receptor binding domain (UNP residues 330-529); light chain of human monoclonal antibody Fab AZD1061; heavy chain of human monoclonal antibody Fab AZD1061; heavy chain of human monoclonal antibody AZD8895 | 45.1 | - | No | - | 18.0 | 88.1 | 59.5 | - | ||||||
| 7r7h | 2.15 Å | 2021-09-01 | X-ray | 4IT | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.31% | 25.92% | N/A | N-[(2S)-1-({(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | N-[(2S)-1-({(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | XDS | 96.64% | 9.23 | SSRL (BL12-1) | PHENIX | 2021-06-24 | 34778773 | - | - | - | - | N/A | - | - | 3C-like proteinase | 20.8 | - | No | - | 27.2 | 48.9 | 40.1 | - | ||||||
| 7rw2 | 3.50 Å | 2021-09-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-08-19 | 34706271 | - | - | - | - | N/A | - | - | 5-7 heavy chain; 5-7 light chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7e5o | 2.80 Å | 2021-09-08 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 22.62% | 25.62% | 20.54% | - | XDS | 99.88% | 5.75 | Photon Factory (BL-17A) | PHENIX | 2021-02-19 | 34508662 | - | - | - | - | N/A | - | - | NT-193 Heavy chain; Spike protein S1; NT-193 Light chain | 45.8 | - | No | - | 29.7 | 69.4 | 68.0 | - | |||||||
| 7ey0 | 3.20 Å | 2021-09-08 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-05-29 | 34433900 | - | - | - | - | N/A | - | - | BD-813L; BD-813H; BD-744H; Spike glycoprotein; BD-744L | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ey4 | 3.69 Å | 2021-09-08 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-05-29 | 34433900 | - | - | - | - | N/A | - | - | BD-667 L; Spike glycoprotein; Spike glycoprotein; BD-667 H | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ey5 | 3.40 Å | 2021-09-08 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | 2021-05-29 | 34433900 | - | - | - | - | N/A | - | - | BD-821L; BD-771H; BD-771L; Spike glycoprotein; BD-821H | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7eya | 3.77 Å | 2021-09-08 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-05-30 | 34433900 | - | - | - | - | N/A | - | - | BD-804H; Spike glycoprotein; BD-804L; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ezv | 3.30 Å | 2021-09-08 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-06-02 | 34433900 | - | - | - | - | N/A | - | - | 836H; Spike glycoprotein; 812 H; 812L; 836L | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7n7r | 2.01 Å | 2021-09-08 | X-ray | S6V | SARS-CoV-2 | NSP15 | Functional ligand | 21.70% | 23.97% | N/A | 1-[2-(2-oxidanylidenepyrrolidin-1-yl)ethyl]-3-phenyl-urea | 1-[2-(2-oxidanylidenepyrrolidin-1-yl)ethyl]-3-phenyl-urea | XDS | 100.00% | 7.1 | Diamond (I04-1) | PHENIX | 2021-06-11 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 53.2 | - | No | - | 45.2 | 60.5 | 75.5 | - | ||||||
| 7n7u | 2.06 Å | 2021-09-08 | X-ray | 0MI | SARS-CoV-2 | NSP15 | Functional ligand | 22.16% | 25.43% | 18.80% | 1-[(2~{R},4~{S},5~{R})-5-[[(azanylidene-$l^{4}-azanylidene)amino]methyl]-4-oxidanyl-oxolan-2-yl]-5-methyl-pyrimidine-2,4-dione | XDS | 94.30% | 8.5 | Diamond (I04-1) | PHENIX | 2021-06-11 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 44.4 | - | No | - | 31.4 | 83.2 | 49.9 | - | |||||||
| 7n7w | 2.42 Å | 2021-09-08 | X-ray | 0OI | SARS-CoV-2 | NSP15 | Functional ligand | 19.38% | 22.81% | 75.10% | N-(2-fluorophenyl)-N'-methylurea | N-(2-fluorophenyl)-N'-methylurea | XDS | 100.00% | 4.0 | Diamond (I04-1) | PHENIX | 2021-06-11 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 68.9 | - | No | - | 56.9 | 90.0 | 66.4 | - | ||||||
| 7n7y | 2.09 Å | 2021-09-08 | X-ray | RZG | SARS-CoV-2 | NSP15 | Functional ligand | 21.70% | 24.08% | 25.60% | methyl 4-sulfamoylbenzoate | methyl 4-sulfamoylbenzoate | XDS | 100.00% | 6.7 | Diamond (I04-1) | PHENIX | 2021-06-11 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 54.6 | - | No | - | 44.2 | 58.6 | 81.0 | - | ||||||
| 7n83 | 1.91 Å | 2021-09-08 | X-ray | WNM | SARS-CoV-2 | NSP15 | Functional ligand | 20.84% | 23.17% | 15.20% | (3S)-1-(phenylsulfonyl)pyrrolidin-3-amine | (3S)-1-(phenylsulfonyl)pyrrolidin-3-amine | XDS | 100.00% | 9.5 | Diamond (I04-1) | BUSTER | 2021-06-12 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | 66.4 | - | No | - | 53.4 | 70.3 | 84.2 | - | ||||||
| 7rn1 | 2.30 Å | 2021-09-08 | X-ray | 5ZF | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.80% | 22.25% | 8.80% | N-([1,1'-biphenyl]-4-yl)-2-chloro-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]acetamide | N-([1,1'-biphenyl]-4-yl)-2-chloro-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]acetamide | iMOSFLM | 76.70% | 7.2 | APS (22-ID) | REFMAC | 2021-07-28 | 34860011 | - | - | - | - | N/A | - | - | 3C-like proteinase | 66.7 | - | No | - | 62.7 | 83.7 | 62.1 | - | ||||||
| 7rzc | 2.04 Å | 2021-09-08 | X-ray | JWX | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 17.76% | 21.17% | 10.90% | (1R)-N-[(1H-indol-3-yl)methyl]-N-methyl-1-(naphthalen-1-yl)ethan-1-amine | (1R)-N-[(1H-indol-3-yl)methyl]-N-methyl-1-(naphthalen-1-yl)ethan-1-amine | HKL-3000 | 100.00% | 6.3 | APS (19-ID) | REFMAC | 2021-08-27 | - | ZN | - | - | - | N/A | - | - | Non-structural protein 3 | 63.0 | - | No | - | - | 72.7 | 53.2 | 74.8 | - | |||||
| 7cyv | 3.13 Å | 2021-09-15 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 25.18% | 27.62% | 27.24% | - | XDS | 98.89% | 5.65 | SSRF (BL18U1) | PHENIX | 2020-09-04 | 34672091 | - | - | - | - | N/A | - | - | Spike protein S1 - receptor-binding domain (RBD); The heavy chain variable region of the scFv FD20,The light chain variable region of the scFv FD20 | 22.7 | - | No | - | 16.0 | 77.3 | 27.1 | - | |||||||
| 7e3k | 3.90 Å | 2021-09-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-02-09 | 34728625 | - | - | - | - | N/A | - | - | 13G9 heavy chain; Spike glycoprotein; 13G9 light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7e3l | 3.60 Å | 2021-09-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-02-09 | 34728625 | - | - | - | - | N/A | - | - | 58G6 heavy chain; Spike glycoprotein; 58G6 light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7e3o | 2.51 Å | 2021-09-15 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 20.40% | 25.89% | N/A | - | HKL-3000 | 94.20% | -0.6 | SSRF (BL19U1) | PHENIX | 2021-02-09 | 34643438 | - | - | - | - | N/A | - | - | nCoV617 Heigh Chain; nCoV617 Light Chain; Spike protein S1 - UNP residues 337-527 | 36.7 | - | No | - | 27.6 | 73.8 | 48.2 | - | |||||||
| 7lq7 | 3.40 Å | 2021-09-15 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 19.74% | 23.34% | 10.20% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 98.80% | 5.7 | SSRL (BL12-1) | REFMAC | 2021-02-13 | 34519517 | - | - | - | - | N/A | - | - | Spike protein S1; COVA1-16 light chain; COVA1-16 heavy chain; CV503 light chain; CV503 heavy chain | 53.7 | - | No | - | 51.6 | 87.3 | 43.2 | - | ||||||
| 7mtc | 2.60 Å | 2021-09-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-05-13 | 34461095 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7mtd | 3.50 Å | 2021-09-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-05-13 | 34461095 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7mte | 3.20 Å | 2021-09-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-05-13 | 34461095 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7s3k | 1.90 Å | 2021-09-15 | X-ray | Z26 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.51% | 20.39% | 7.80% | 2-(5-chloro-2-methoxyphenyl)-N-(isoquinolin-4-yl)acetamide | 2-(5-chloro-2-methoxyphenyl)-N-(isoquinolin-4-yl)acetamide | CrysalisPro | 99.90% | 13.39 | ROTATING ANODE () | PHENIX | 2021-09-07 | 35434531 | - | - | - | - | N/A | - | - | 3C-like proteinase | 71.4 | - | No | - | 79.0 | 42.8 | 97.1 | - | ||||||
| 7s3s | 2.00 Å | 2021-09-15 | X-ray | 860 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.32% | 22.20% | 5.90% | 2-(3-chlorophenyl)-N-(isoquinolin-4-yl)acetamide | 2-(3-chlorophenyl)-N-(isoquinolin-4-yl)acetamide | CrysalisPro | 98.60% | 18.4 | ROTATING ANODE () | PHENIX | 2021-09-08 | 35434531 | - | - | - | - | N/A | - | - | 3C-like proteinase | 68.8 | - | No | - | 63.1 | 52.3 | 97.6 | - | ||||||
| 7s4b | 2.00 Å | 2021-09-15 | X-ray | 87H | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.17% | 20.98% | 7.80% | (2R)-2-(3-fluorophenyl)-N-(isoquinolin-4-yl)propanamide | (2R)-2-(3-fluorophenyl)-N-(isoquinolin-4-yl)propanamide | CrysalisPro | 97.50% | 12.51 | ROTATING ANODE () | PHENIX | 2021-09-08 | 35434531 | - | - | - | - | N/A | - | - | 3C-like proteinase | 75.3 | - | No | - | 74.3 | 56.1 | 98.2 | - | ||||||
| 7eiz | 3.35 Å | 2021-09-22 | Cryo-EM | - | SARS-CoV-2 | NSP7/NSP8/NSP9/NSP10/NSP12/NSP13/NSP14 | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-04-01 | 34143953 | ZN; MG | - | - | - | N/A | - | - | Helicase - UNP residues 5325-5925; Non-structural protein 8 - UNP residues 3943-4140; RNA-directed RNA polymerase - UNP residues 4393-5324; Non-structural protein 7 - UNP residues 3860-3942; Non-structural protein 10 - UNP residues 4254-4392; primer; Proofreading exoribonuclease - UNP residues 5926-6452; template; Non-structural protein 9 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7kn3 | 2.25 Å | 2021-09-22 | X-ray | NAG, PGE | SARS-CoV-2 | Spike | Pathogen-host interaction | 18.63% | 22.19% | 9.80% | 2-acetamido-2-deoxy-beta-D-glucopyranose; TRIETHYLENE GLYCOL | 2-acetamido-2-deoxy-beta-D-glucopyranose; TRIETHYLENE GLYCOL | HKL-2000 | 97.40% | 6.0 | APS (23-ID-B) | PHENIX | 2020-11-04 | 34716683 | - | - | - | - | N/A | - | - | S-B8 Fab light chain; Spike protein S1; S-B8 Fab heavy chain | 72.1 | - | No | - | 63.4 | 69.8 | 87.4 | - | ||||||
| 7kn4 | 2.70 Å | 2021-09-22 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 24.08% | 29.10% | 9.20% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 93.30% | 7.7 | APS (23-ID-B) | PHENIX | 2020-11-04 | 34716683 | - | - | - | - | N/A | - | - | S-E6 Fab heavy chain; Spike protein S1; S-E6 Fab light chain | 17.1 | - | No | - | 9.6 | 45.9 | 52.3 | - | ||||||
| 7l8i | 2.10 Å | 2021-09-22 | X-ray | AG7 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.63% | 26.31% | 8.65% | 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | HKL-3000 | 92.20% | 9.05 | ROTATING ANODE () | PHENIX | 2020-12-31 | 34506130 | - | - | - | - | N/A | - | - | 3C-like proteinase | 22.7 | - | No | - | 24.3 | 26.5 | 69.7 | - | ||||||
| 7l8j | 2.45 Å | 2021-09-22 | X-ray | AG7 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 22.33% | 28.11% | 12.47% | 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | 99.72% | 10.04 | NSLS-II (17-ID-1) | PHENIX | 2020-12-31 | 34506130 | - | - | - | - | N/A | - | - | 3C-like proteinase | 11.3 | - | No | - | 13.5 | 15.6 | 63.5 | - | |||||||
| 7rks | 2.70 Å | 2021-09-22 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 19.46% | 24.67% | 6.22% | - | XDS | 95.70% | 13.14 | SSRL (BL12-2) | PHENIX | 2021-07-22 | 34534459 | - | - | - | - | N/A | - | - | Spike protein S1; C118 Antibody Fab Light Chain; C118 Antibody Fab Heavy Chain | 36.8 | - | No | - | 38.5 | 65.9 | 45.2 | - | |||||||
| 7rku | 3.20 Å | 2021-09-22 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 18.76% | 23.11% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.50% | 12.87 | SSRL (BL12-2) | PHENIX | 2021-07-22 | 34534459 | - | - | - | - | N/A | - | - | C022 Antibody Fab Heavy Chain; C022 Antibody Fab Light Chain; Spike protein S1 - Receptor Binding Domain | 37.8 | - | No | - | 53.9 | 60.5 | 37.3 | - | ||||||
| 7rkv | 3.45 Å | 2021-09-22 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-07-22 | 34534459 | - | - | - | - | N/A | - | - | Spike glycoprotein; C118 Fab Light Chain; C118 Fab Heavy Chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7s4s | 2.05 Å | 2021-09-22 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 16.57% | 19.79% | 14.10% | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-3000 | 98.00% | 11.6 | SSRL (BL12-2) | REFMAC; PHENIX | 2021-09-09 | 37334379 | - | - | - | - | N/A | - | - | Spike protein S1; CoV11 heavy chain; CoV11 light chain | 53.1 | - | No | - | 83.2 | 26.0 | 71.9 | - | |||||||
| 7d64 | 2.45 Å | 2021-09-29 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.63% | 26.54% | 7.80% | - | XDS | 99.00% | 10.3 | CLSI (08ID-1) | REFMAC | 2020-09-29 | 35255411 | ZN | - | - | - | N/A | - | - | 3C-like proteinase | 31.7 | - | No | - | 22.7 | 73.2 | 43.4 | - | |||||||
| 7dhx | 2.30 Å | 2021-09-29 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 19.44% | 23.49% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.20% | 51.4 | SSRF (BL17U) | PHENIX | 2020-11-17 | - | ZN | - | - | - | N/A | - | - | Spike glycoprotein - Receptor binding domain (RBD); pangolin ACE2 | 44.1 | - | No | - | - | 50.2 | 56.4 | 57.2 | - | ||||||
| 7dqz | 1.99 Å | 2021-09-29 | X-ray | - | SARS-CoV | NSP5 (3CLpro) | No functional ligands | 22.83% | 25.75% | 3.20% | - | AUTOMAR | 94.10% | 2.0 | SSRF (BL17U1) | PHENIX | 2020-12-24 | 34586857 | - | - | - | - | N/A | - | - | 3C-like proteinase | 18.0 | - | No | - | 28.7 | 19.8 | 61.5 | - | |||||||
| 7fg2 | 4.40 Å | 2021-09-29 | Cryo-EM | - | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | - | - | - | () | 2021-07-25 | 34648602 | - | - | - | - | N/A | - | - | K-874A VHH; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7fg3 | 3.90 Å | 2021-09-29 | Cryo-EM | - | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | - | - | - | () | 2021-07-25 | 34648602 | - | - | - | - | N/A | - | - | K-874A VHH; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7fg7 | 6.90 Å | 2021-09-29 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2021-07-26 | 34648602 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7k9h | 3.20 Å | 2021-09-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2020-09-29 | 34655519 | - | - | - | - | N/A | - | - | Spike glycoprotein; 2B04 heavy chain; 2B04 light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7k9i | 3.30 Å | 2021-09-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-09-29 | 34655519 | - | - | - | - | N/A | - | - | 2B04 heavy chain; Spike protein S1 - receptor binding domain (UNP residues 333-527); 2B04 light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7k9j | 3.00 Å | 2021-09-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2020-09-29 | 34655519 | - | - | - | - | N/A | - | - | Spike glycoprotein; 2H04 heavy chain; 2H04 light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7k9k | 3.14 Å | 2021-09-29 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | phenix.real_space_refine; PHENIX | 2020-09-29 | 34655519 | - | - | - | - | N/A | - | - | Spike protein S1 - receptor binding domain (UNP residues 333-527); 2H04 heavy chain; 2H04 light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7rbr | 1.88 Å | 2021-09-29 | X-ray | - | SARS-CoV-2 | NSP3: PLpro | Protein-protein complex | 18.67% | 22.84% | 10.60% | - | HKL-3000 | 96.10% | 6.5 | APS (19-ID) | REFMAC | 2021-07-06 | 37185902 | ZN | - | - | - | N/A | - | - | Papain-like protease; Ubiquitin | 54.9 | - | No | - | 56.6 | 44.0 | 83.8 | - | |||||||
| 7rbs | 2.98 Å | 2021-09-29 | X-ray | - | SARS-CoV-2 | NSP3: PLpro | Protein-protein complex | 19.51% | 23.58% | 21.40% | - | HKL-3000 | 99.90% | 5.3 | APS (19-ID) | REFMAC | 2021-07-06 | 37185902 | ZN | - | - | - | N/A | - | - | Papain-like protease; Ubiquitin-like protein ISG15 | 70.7 | - | No | - | 49.2 | 83.0 | 85.1 | - | |||||||
| 7rbz | 1.65 Å | 2021-09-29 | X-ray | 4IJ | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.72% | 18.92% | 7.79% | 5-chloropyridin-3-yl 2,3-dihydro-1H-indole-4-carboxylate | 5-chloropyridin-3-yl 2,3-dihydro-1H-indole-4-carboxylate | HKL-2000 | 99.79% | 16.18 | APS (21-ID-F) | REFMAC; PHENIX | 2021-07-06 | 34528437 | - | - | - | - | N/A | - | - | 3C-like proteinase | 52.0 | - | No | - | 88.2 | 29.4 | 61.3 | - | ||||||
| 7rc0 | 1.65 Å | 2021-09-29 | X-ray | 4I9, NA | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 12.59% | 17.41% | 7.27% | 5-chloro-4-methylpyridin-3-yl 1H-indole-4-carboxylate; SODIUM ION | 5-chloro-4-methylpyridin-3-yl 1H-indole-4-carboxylate; SODIUM ION | HKL-2000 | 99.53% | 17.86 | APS (21-ID-F) | PHENIX | 2021-07-06 | 34528437 | - | - | - | - | N/A | - | - | 3C-like proteinase | 69.6 | - | No | - | 94.1 | 52.2 | 68.6 | - | ||||||
| 7rc1 | 1.63 Å | 2021-09-29 | X-ray | 4IO | SARS-CoV | NSP5 (3CLpro) | Functional ligand | 19.05% | 22.17% | 5.83% | 5-chloropyridin-3-yl 1-(3-nitrobenzene-1-sulfonyl)-1H-indole-5-carboxylate | 5-chloropyridin-3-yl 1-(3-nitrobenzene-1-sulfonyl)-1H-indole-5-carboxylate | HKL-2000 | 97.67% | 23.62 | APS (21-ID-G) | PHENIX; PHENIX | 2021-07-07 | 34528437 | - | - | - | - | N/A | - | - | 3C-like proteinase | 31.8 | - | No | - | 63.5 | 17.7 | 58.3 | - | ||||||
| 7e6l | 1.78 Å | 2021-10-06 | X-ray | - | HCoV-NL63 | NSP5 (3CLpro) | No functional ligands | 18.10% | 20.16% | 4.60% | - | HKL-2000 | 99.70% | 19.9 | SSRF (BL17U1) | PHENIX | 2021-02-22 | 34605439 | - | - | - | - | N/A | - | - | 3C-like proteinase | 60.3 | - | No | - | 80.6 | 53.3 | 61.1 | - | |||||||
| 7e6m | 1.83 Å | 2021-10-06 | X-ray | - | HCoV-NL63 | NSP5 (3CLpro) | No functional ligands | 19.13% | 22.53% | 6.00% | - | HKL-2000 | 99.90% | 18.5 | SSRF (BL17U1) | PHENIX | 2021-02-22 | 34605439 | - | - | - | - | N/A | - | - | 3C-like proteinase | 47.1 | - | No | - | 59.9 | 53.3 | 56.3 | - | |||||||
| 7e6n | 1.84 Å | 2021-10-06 | X-ray | - | HCoV-NL63 | NSP5 (3CLpro) | No functional ligands | 18.40% | 21.74% | 4.60% | - | HKL-2000 | 99.20% | 20.2 | SSRF (BL17U1) | PHENIX | 2021-02-22 | 34605439 | - | - | - | - | N/A | - | - | 3C-like proteinase | 54.7 | - | No | - | 67.6 | 59.3 | 57.3 | - | |||||||
| 7e6r | 1.90 Å | 2021-10-06 | X-ray | - | HCoV-NL63 | NSP5 (3CLpro) | No functional ligands | 18.70% | 22.24% | 7.50% | - | HKL-2000 | 99.60% | 16.5 | SSRF (BL17U1) | PHENIX | 2021-02-23 | 34605439 | - | - | - | - | N/A | - | - | 3C-like proteinase | 51.1 | - | No | - | 62.8 | 51.5 | 63.0 | - | |||||||
| 7mzf | 2.49 Å | 2021-10-06 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 18.51% | 22.72% | 17.10% | - | XDS | 99.50% | 11.33 | Australian Synchrotron (MX2) | PHENIX | 2021-05-24 | 34610292 | - | - | - | - | N/A | - | - | PDI 37 light chain; PDI 37 heavy chain; Spike protein S1 - Receptor Binding Domain (RBD) | 58.2 | - | No | - | 57.9 | 88.9 | 44.2 | - | |||||||
| 7mzg | 2.00 Å | 2021-10-06 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 20.03% | 23.64% | 10.10% | - | XDS | 99.70% | 11.82 | Australian Synchrotron (MX2) | PHENIX | 2021-05-24 | 34610292 | - | - | - | - | N/A | - | - | PDI 42 heavy chain; PDI 42 light chain; Spike protein S1 - Receptor Binding Domain (RBD) | 39.1 | - | No | - | 48.5 | 25.3 | 80.4 | - | |||||||
| 7mzh | 2.10 Å | 2021-10-06 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 21.99% | 26.53% | 7.30% | - | XDS | 99.90% | 11.8 | Australian Synchrotron (MX2) | PHENIX | 2021-05-24 | 34610292 | - | - | - | - | N/A | - | - | WCSL 119 light chain; Spike protein S1 - Receptor Binding Domain (RBD); WCSL 119 heavy chain | 34.8 | - | No | - | 22.7 | 58.9 | 64.0 | - | |||||||
| 7mzi | 1.85 Å | 2021-10-06 | X-ray | PG4 | SARS-CoV-2 | Spike | Pathogen-host interaction | 17.40% | 20.75% | 7.90% | TETRAETHYLENE GLYCOL | XDS | 99.30% | 9.6 | Australian Synchrotron (MX2) | PHENIX | 2021-05-24 | 34610292 | - | - | - | - | N/A | - | - | WCSL 129 heavy chain; Spike protein S1 - Receptor Binding Domain (RBD); WCSL 129 light chain | 38.9 | - | No | - | 76.2 | 27.3 | 50.3 | - | |||||||
| 7mzj | 2.40 Å | 2021-10-06 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 22.21% | 26.82% | 9.60% | - | XDS | 98.40% | 9.28 | Australian Synchrotron (MX2) | PHENIX | 2021-05-24 | 34610292 | - | - | - | - | N/A | - | - | WCSL 129 light chain; PDI 93 heavy chain; PDI 93 light chain; Spike protein S1 - Receptor Binding Domain (RBD); WCSL 129 heavy chain | 25.9 | - | No | - | 20.7 | 69.3 | 37.3 | - | |||||||
| 7mzk | 2.25 Å | 2021-10-06 | X-ray | MPD | SARS-CoV-2 | Spike | Pathogen-host interaction | 19.72% | 23.51% | 10.80% | (4S)-2-METHYL-2,4-PENTANEDIOL | (4S)-2-METHYL-2,4-PENTANEDIOL | XDS | 98.20% | 8.13 | Australian Synchrotron (MX2) | PHENIX | 2021-05-24 | 34610292 | - | - | - | - | N/A | - | - | PDI 96 heavy chain; WCSL 129 light chain; Spike protein S1 - Receptor Binding Domain (RBD); WCSL 129 heavy chain; PDI 96 light chain | 31.9 | - | No | - | 49.8 | 49.6 | 40.3 | - | ||||||
| 7mzl | 3.70 Å | 2021-10-06 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 22.15% | 25.62% | 34.40% | - | XDS | 99.90% | 7.5 | Australian Synchrotron (MX2) | PHENIX | 2021-05-24 | 34610292 | - | - | - | - | N/A | - | - | PDI 210 heavy chain; Spike protein S1 - Receptor Binding Domain (RBD); PDI 210 light chain | 60.8 | - | No | - | 29.7 | 91.2 | 75.1 | - | |||||||
| 7mzm | 2.30 Å | 2021-10-06 | X-ray | IPA | SARS-CoV-2 | Spike | Pathogen-host interaction | 21.60% | 25.73% | 16.40% | ISOPROPYL ALCOHOL | ISOPROPYL ALCOHOL | XDS | 99.80% | 11.58 | Australian Synchrotron (MX2) | PHENIX | 2021-05-24 | 34610292 | - | - | - | - | N/A | - | - | PDI 215 light chain; PDI 215 heavy chain; Spike protein S1 - Receptor Binding Domain (RBD) | 23.0 | - | No | - | 28.8 | 47.2 | 45.2 | - | ||||||
| 7mzn | 3.10 Å | 2021-10-06 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 22.33% | 25.85% | 27.90% | - | XDS | 99.90% | 6.5 | Australian Synchrotron (MX2) | PHENIX | 2021-05-24 | 34610292 | - | - | - | - | N/A | - | - | PDI 231 light chain; PDI 231 heavy chain; Spike protein S1 - Receptor Binding Domain (RBD) | 61.7 | - | No | - | 27.8 | 89.1 | 81.1 | - | |||||||
| 7n4i | 2.28 Å | 2021-10-06 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 19.99% | 23.78% | N/A | - | HKL-2000 | 99.30% | 19.0 | APS (24-ID-C) | PHENIX | 2021-06-04 | 34716452 | - | - | - | - | N/A | - | - | WRAIR-2057 Antibody Fab Light Chain; Spike protein S1 - receptor binding domain (RBD); WRAIR-2057 Antibody Fab Heavy Chain | 21.4 | - | No | - | 47.2 | 15.2 | 55.3 | - | |||||||
| 7n4j | 2.21 Å | 2021-10-06 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 17.20% | 20.69% | N/A | - | HKL-2000 | 99.20% | 10.6 | APS (19-ID) | PHENIX | 2021-06-04 | 34716452 | - | - | - | - | N/A | - | - | Spike protein S1 - Receptor Binding Domain (RBD); WRAIR-2173 antibody Fab light chain; WRAIR-2173 antibody Fab heavy chain | 45.4 | - | No | - | 76.7 | 24.1 | 65.4 | - | |||||||
| 7nts | 1.48 Å | 2021-10-06 | X-ray | FMT | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.57% | 20.72% | N/A | FORMIC ACID | FORMIC ACID | Aimless | 97.10% | 11.7 | SOLEIL (PROXIMA 1) | REFMAC | 2021-03-10 | 34570415 | - | - | - | - | N/A | - | - | Replicase polyprotein 1ab | 76.9 | - | No | - | 76.3 | 73.6 | 83.0 | - | ||||||
| 7p51 | 1.47 Å | 2021-10-06 | X-ray | 5P9, NA | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.14% | 21.57% | N/A | SODIUM ION; N-(5-chloropyridin-2-yl)-3-oxo-2,3-dihydro-1H-indene-1-carboxamide | N-(5-chloropyridin-2-yl)-3-oxo-2,3-dihydro-1H-indene-1-carboxamide; SODIUM ION | XDS | 99.40% | 13.68 | SOLEIL (PROXIMA 2) | REFMAC | 2021-07-13 | 34570415 | - | - | - | - | N/A | - | - | 3C-like proteinase | 62.6 | - | No | - | 69.2 | 55.9 | 74.8 | - | ||||||
| 7rr0 | 3.12 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-08-08 | 34610292 | - | - | - | - | N/A | - | - | PDI 222 Fab Heavy Chain; Spike protein S1 - RBD domain; PDI 222 Fab Light Chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7rxd | 3.60 Å | 2021-10-06 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-08-22 | 34620716 | - | - | - | - | N/A | - | - | Fab_8D3_2 light chain; Spike protein S1 - Receptor Binding Domain (RBD); Maltodextrin-binding protein,Immunoglobulin G-binding protein A,Immunoglobulin G-binding protein G; Fab_8D3_2 heavy chain; Nb_RBD | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7s0b | 2.90 Å | 2021-10-06 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 21.46% | 25.97% | 18.70% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.80% | 6.4 | SSRL (BL12-2) | PHENIX | 2021-08-30 | 35114110 | - | - | - | - | N/A | - | - | N-612-056 Light Chain; Spike protein S1; N-612-056 Fab Heavy Chain | 7.2 | - | No | - | 26.9 | 22.7 | 29.4 | - | ||||||
| 7s0c | 3.25 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-08-30 | 35114110 | - | - | - | - | N/A | - | - | Spike glycoprotein; N-612-017 Fab Heavy Chain; N-612-017 Light Chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7s0d | 3.50 Å | 2021-10-06 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-08-30 | 35114110 | - | - | - | - | N/A | - | - | N-612-014 Light Chain; Spike glycoprotein; N-612-014 Fab Heavy Chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7s0e | 4.90 Å | 2021-10-06 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-08-30 | 35114110 | - | - | - | - | N/A | - | - | N-612-004 Fab heavy chain; Spike glycoprotein; N-612-004 Light Chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7sdr | 2.72 Å | 2021-10-06 | X-ray | JW9 | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 17.29% | 21.22% | 14.00% | 4-({methyl[(1R)-1-(naphthalen-1-yl)ethyl]amino}methyl)phenol | 4-({methyl[(1R)-1-(naphthalen-1-yl)ethyl]amino}methyl)phenol | HKL-3000 | 99.80% | 5.8 | APS (19-ID) | REFMAC | 2021-09-29 | - | ZN | - | - | - | N/A | - | - | Non-structural protein 3 | 77.5 | - | No | - | - | 72.2 | 77.6 | 84.9 | - | |||||
| 7v76 | 3.20 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-08-21 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v77 | 3.30 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-08-21 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v78 | 3.40 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-08-21 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v79 | 3.30 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-08-21 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v7a | 3.40 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-08-21 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v7d | 3.00 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-08-21 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v7e | 2.90 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-08-21 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v7f | 2.90 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-08-21 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v7g | 3.10 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-08-21 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v7h | 3.20 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-08-21 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v7i | 3.30 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-08-21 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v7j | 3.40 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-08-21 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v7n | 2.90 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-08-21 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v7o | 2.90 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-08-21 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v7p | 2.90 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-08-21 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v7q | 2.80 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-08-21 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v7r | 2.90 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-08-21 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v7s | 3.00 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-08-21 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v7t | 3.00 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-08-21 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v7u | 3.00 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-08-21 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v7v | 3.10 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-08-21 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v7z | 3.10 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-08-22 | - | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2,Green fluorescent protein; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v80 | 3.90 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-08-22 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2,Green fluorescent protein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v81 | 3.20 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-08-22 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2,Green fluorescent protein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v82 | 2.80 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-08-22 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2,Green fluorescent protein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v83 | 2.80 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-08-22 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2,Green fluorescent protein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v84 | 3.00 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-08-22 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2,Green fluorescent protein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v85 | 3.30 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-08-22 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2,Green fluorescent protein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v86 | 2.80 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-08-22 | - | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2,Green fluorescent protein; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v87 | 3.30 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-08-22 | - | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2,Angiotensin-converting enzyme 2 (ACE2) ectodomain; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v88 | 3.30 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-08-22 | - | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2,Angiotensin-converting enzyme 2 (ACE2) ectodomain; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v89 | 2.80 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-08-22 | - | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2,Angiotensin-converting enzyme 2 (ACE2) ectodomain; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v8a | 2.70 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-08-22 | - | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2,Angiotensin-converting enzyme 2 (ACE2) ectodomain; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v8b | 3.20 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-08-22 | - | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2,Angiotensin-converting enzyme 2 (ACE2) ectodomain; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7v8c | 3.40 Å | 2021-10-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-08-22 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7d6i | 3.41 Å | 2021-10-13 | X-ray | - | SARS-CoV | Spike | Pathogen-host interaction | 22.55% | 25.51% | N/A | - | HKL-2000 | 100.00% | 12.6 | SSRF (BL17U1) | PHENIX | 2020-09-30 | - | - | - | - | - | N/A | - | - | Heavy chain of GH12-Fab; SARS-CoV-2 receptor binding domain; Light chain of GH12-Fab | 27.4 | - | No | - | - | 30.6 | 65.2 | 34.5 | - | ||||||
| 7e5r | 3.60 Å | 2021-10-13 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-02-20 | 33782529 | - | - | - | - | N/A | - | - | FC05 heavy chain; H014 light chain; H014 heavy chain; FC05 light chain; P17 light chain; P17 heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7jp0 | 1.65 Å | 2021-10-13 | X-ray | VJA | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.83% | 22.60% | N/A | N-[(benzyloxy)carbonyl]-L-valyl-N-{(2R)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | N-[(benzyloxy)carbonyl]-L-valyl-N-{(2R)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | MOSFLM | 96.60% | 8.9 | ALS (5.0.2) | PHENIX | 2020-08-07 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 31.5 | - | No | - | - | 59.1 | 29.3 | 50.5 | - | |||||
| 7p40 | 3.50 Å | 2021-10-13 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | 2021-07-09 | 34599871 | - | - | - | - | N/A | - | - | Spike glycoprotein; Variable Light Chain P5C3 (VL); Variable Heavy Chain P5C3 (VH) | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7phg | 4.30 Å | 2021-10-13 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2021-08-17 | 34599871 | - | - | - | - | N/A | - | - | Heavy ChaIn variable; Surface glycoprotein; Light ChaIn | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7dte | 3.00 Å | 2021-10-20 | Cryo-EM | - | SARS-CoV-2 | NSP7/NSP8/NSP12 | Protein-protein complex | N/A | N/A | N/A | RNA (57-MER); RNA (33-MER) | - | - | - | () | PHENIX | 2021-01-04 | 34653416 | ZN | - | - | - | N/A | - | - | Non-structural protein 8; RNA (57-MER); RNA (33-MER); RNA-directed RNA polymerase; Non-structural protein 7 | N/A | - | No | - | N/A | N/A | N/A | RNA (57-MER); RNA (33-MER) | - | ||||||
| 7eo7 | 2.25 Å | 2021-10-20 | X-ray | FNO | HCoV-NL63 | NSP5 (3CLpro) | Functional ligand | 17.65% | 23.21% | 6.70% | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione | HKL-2000 | 99.40% | 16.1 | SSRF (BL17U1) | PHENIX; PHENIX | 2021-04-21 | 34586857 | - | - | - | - | N/A | - | - | 3C-like proteinase | 52.2 | - | No | - | 52.9 | 68.2 | 58.3 | - | ||||||
| 7eo8 | 2.28 Å | 2021-10-20 | X-ray | FNO | SARS-CoV | NSP5 (3CLpro) | Functional ligand | 22.72% | 27.20% | 4.60% | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione | HKL-2000 | 95.00% | 14.4 | SSRF (BL17U1) | PHENIX; PHENIX | 2021-04-21 | 34586857 | - | - | - | - | N/A | - | - | 3C-like proteinase | 6.4 | - | No | - | 18.5 | 17.3 | 40.0 | - | ||||||
| 7f2e | 3.10 Å | 2021-10-20 | X-ray | PO4 | SARS-CoV-2 | Nucleocapsid | No functional ligands | 26.76% | 29.75% | 12.40% | PHOSPHATE ION | PHOSPHATE ION | HKL-3000 | 99.90% | 5.4 | SSRF (BL18U1) | PHENIX | 2021-06-10 | 34665939 | - | - | - | - | N/A | - | - | Nucleoprotein | 36.6 | - | No | - | 7.9 | 86.3 | 55.1 | - | ||||||
| 7orr | 1.79 Å | 2021-10-20 | X-ray | PIM | SARS-CoV-2 | NSP10 | Functional ligand | 17.15% | 18.44% | 4.70% | 4-PHENYL-1H-IMIDAZOLE | 4-PHENYL-1H-IMIDAZOLE | XDS | 99.30% | 24.3 | MAX IV (BioMAX) | REFMAC | 2021-06-06 | 35128408 | ZN | - | - | - | N/A | - | - | Non-structural protein 10 | 66.1 | - | No | - | 90.5 | 18.5 | 98.2 | - | ||||||
| 7oru | 1.67 Å | 2021-10-20 | X-ray | 2AQ | SARS-CoV-2 | NSP10 | Functional ligand | 16.05% | 17.17% | 7.10% | QUINOLIN-2-AMINE | QUINOLIN-2-AMINE | XDS | 100.00% | 27.3 | MAX IV (BioMAX) | REFMAC | 2021-06-06 | 35128408 | ZN | - | - | - | N/A | - | - | Non-structural protein 10 | 71.5 | - | No | - | 94.7 | 25.0 | 99.3 | - | ||||||
| 7orv | 1.95 Å | 2021-10-20 | X-ray | X4V | SARS-CoV-2 | NSP10 | Functional ligand | 17.03% | 20.20% | 7.20% | N~4~,N~4~-dimethylpyridine-2,4-diamine | N~4~,N~4~-dimethylpyridine-2,4-diamine | XDS | 99.80% | 14.8 | MAX IV (BioMAX) | REFMAC | 2021-06-06 | 35128408 | ZN | - | - | - | N/A | - | - | Non-structural protein 10 | 56.4 | - | No | - | 80.3 | 16.1 | 91.0 | - | ||||||
| 7orw | 1.95 Å | 2021-10-20 | X-ray | 7WA | SARS-CoV-2 | NSP10 | Functional ligand | 17.45% | 20.86% | 7.50% | 1H-benzimidazol-4-amine | 1H-benzimidazol-4-amine | XDS | 100.00% | 13.5 | MAX IV (BioMAX) | REFMAC | 2021-06-06 | 35128408 | ZN | - | - | - | N/A | - | - | Non-structural protein 10 | 71.5 | - | No | - | 75.2 | 44.7 | 99.3 | - | ||||||
| 7r95 | - | 2021-10-20 | NMR | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2021-06-28 | 34623907 | - | - | - | - | N/A | - | - | Spike protein S2 - Heptad repeat 1 domain, residues 919-965 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7sgu | 1.79 Å | 2021-10-20 | X-ray | 9EI, FMT | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 16.71% | 18.77% | 11.10% | 5-amino-N-(naphthalen-1-yl)pyridine-3-carboxamide; FORMIC ACID | 5-amino-N-(naphthalen-1-yl)pyridine-3-carboxamide; FORMIC ACID | HKL-3000 | 100.00% | 6.9 | APS (19-ID) | REFMAC | 2021-10-07 | - | ZN | - | - | - | N/A | - | - | Papain-like protease | 79.6 | - | No | - | - | 89.0 | 56.8 | 95.8 | - | |||||
| 7sgv | 2.00 Å | 2021-10-20 | X-ray | L30 | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 18.06% | 20.74% | 18.90% | N-(naphthalen-1-yl)pyridine-3-carboxamide | N-(naphthalen-1-yl)pyridine-3-carboxamide | HKL-3000 | 99.00% | 7.0 | APS (19-ID) | REFMAC | 2021-10-07 | - | ZN | - | - | - | N/A | - | - | Papain-like protease | 54.6 | - | No | - | - | 76.2 | 21.3 | 86.4 | - | |||||
| 7sgw | 1.95 Å | 2021-10-20 | X-ray | L30 | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 17.64% | 20.68% | 12.60% | N-(naphthalen-1-yl)pyridine-3-carboxamide | N-(naphthalen-1-yl)pyridine-3-carboxamide | HKL-3000 | 99.80% | 10.0 | APS (19-BM) | REFMAC | 2021-10-07 | - | ZN | - | - | - | N/A | - | - | Non-structural protein 3 | 60.5 | - | No | - | - | 76.8 | 28.7 | 90.1 | - | |||||
| 7si9 | 2.00 Å | 2021-10-20 | X-ray | 9I7 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.16% | 20.67% | 8.70% | (1R,2S,5S)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | CrysalisPro | 97.70% | 14.5 | ROTATING ANODE () | PHENIX | 2021-10-12 | 35477935 | - | - | - | - | N/A | - | - | 3C-like proteinase | 60.5 | - | No | - | 76.8 | 25.3 | 93.4 | - | ||||||
| 7v26 | 3.85 Å | 2021-10-20 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-08-07 | 34554412 | - | - | - | - | N/A | - | - | Spike glycoprotein; XG005 Heavy chain; XG005 Light Chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7v2a | 3.40 Å | 2021-10-20 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-08-07 | 34554412 | - | - | - | - | N/A | - | - | The heavy chain of XG014; Spike glycoprotein; The light chain of XG014 Fab | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7n3k | 3.00 Å | 2021-10-27 | X-ray | ODN | SARS-CoV-2 | NSP9 | Functional ligand | 22.63% | 28.55% | N/A | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one | MOSFLM | 99.10% | 10.5 | Australian Synchrotron (MX2) | PHENIX | 2021-06-01 | 34756886 | - | - | - | - | N/A | - | - | Non-structural protein 9 | 25.2 | - | No | - | 11.7 | 87.9 | 26.2 | - | ||||||
| 7vnu | 1.95 Å | 2021-10-27 | X-ray | - | SARS-CoV-2 | Nucleocapsid | No functional ligands | 18.84% | 22.59% | 5.00% | - | XDS | 99.80% | 19.8 | SSRF (BL19U1) | REFMAC | 2021-10-12 | 36806252 | - | - | - | - | N/A | - | - | Nucleoprotein - N-terminal domain | 47.7 | - | No | - | 59.4 | 28.1 | 83.1 | - | |||||||
| 7d3c | 2.20 Å | 2021-11-03 | X-ray | - | MERS-CoV | NSP5 (3CLpro) | No functional ligands | 22.16% | 27.80% | N/A | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | - | 100.00% | 6.0 | SSRF (BL17U) | PHENIX | 2020-09-18 | 23549610 | - | - | - | - | N/A | - | - | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE; 3C-like proteinase | 13.8 | - | No | - | 15.0 | 68.7 | 15.7 | - | |||||||
| 7dat | 2.75 Å | 2021-11-03 | X-ray | AU | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 19.41% | 22.83% | 9.40% | GOLD ION | XDS | 96.40% | 14.0 | SSRF (BL17U1) | PHENIX; REFMAC | 2020-10-18 | - | AU | - | - | - | N/A | - | - | COVID-19 MAIN PROTEASE | 64.3 | - | No | - | - | 56.7 | 68.5 | 78.3 | - | ||||||
| 7dau | 1.72 Å | 2021-11-03 | X-ray | AU | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 19.92% | 23.75% | 8.90% | GOLD ION | HKL-3000 | 99.80% | 33.8 | SSRF (BL18U1) | REFMAC; PHENIX | 2020-10-18 | - | AU | - | - | - | N/A | - | - | COVID-19 MAIN PROTEASE | 33.1 | - | No | - | - | 47.4 | 20.8 | 74.0 | - | ||||||
| 7dav | 1.77 Å | 2021-11-03 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 20.65% | 24.75% | 7.00% | - | HKL-3000 | 99.30% | 24.3 | SSRF (BL19U1) | PHENIX; REFMAC | 2020-10-18 | - | - | - | - | - | N/A | - | - | COVID-19 MAIN PROTEASE | 40.0 | - | No | - | - | 37.7 | 51.8 | 66.4 | - | ||||||
| 7ekc | 2.80 Å | 2021-11-03 | X-ray | NAG | SARS-CoV-2 | Spike | No functional ligands | 21.41% | 22.52% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | 99.90% | 17.884 | SSRF (BL17U1) | PHENIX | 2021-04-05 | 34671049 | ZN | - | - | - | N/A | - | - | Spike protein S1; Angiotensin-converting enzyme 2 | 27.5 | - | No | - | 60.0 | 21.5 | 49.1 | - | |||||||
| 7eke | 2.70 Å | 2021-11-03 | X-ray | NAG | SARS-CoV-2 | Spike | No functional ligands | 21.19% | 24.07% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | 100.00% | 20.354 | SSRF (BL17U1) | PHENIX | 2021-04-05 | 34671049 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike protein S1 | 27.7 | - | No | - | 44.3 | 22.7 | 64.1 | - | |||||||
| 7ekf | 2.85 Å | 2021-11-03 | X-ray | NAG | SARS-CoV-2 | Spike | No functional ligands | 20.81% | 22.38% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | 99.90% | 16.5 | SSRF (BL17U1) | PHENIX | 2021-04-05 | 34671049 | ZN | - | - | - | N/A | - | - | Spike protein S1; Angiotensin-converting enzyme 2 | 39.5 | - | No | - | 61.5 | 41.3 | 52.2 | - | |||||||
| 7ekg | 2.63 Å | 2021-11-03 | X-ray | NAG | SARS-CoV-2 | Spike | No functional ligands | 21.38% | 24.84% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | 99.00% | 21.9 | SSRF (BL17U1) | PHENIX | 2021-04-05 | 34671049 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike protein S1 | 24.0 | - | No | - | 36.7 | 27.3 | 59.4 | - | |||||||
| 7ekh | 2.40 Å | 2021-11-03 | X-ray | NAG | SARS-CoV-2 | Spike | No functional ligands | 20.98% | 22.80% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | 99.80% | 19.826 | SSRF (BL17U1) | PHENIX | 2021-04-05 | 34671049 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike protein S1 | 41.5 | - | No | - | 57.0 | 28.4 | 73.6 | - | |||||||
| 7n5h | 3.24 Å | 2021-11-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-06-05 | 34836485 | - | - | - | - | N/A | - | - | 2-36 Fab light chain; Spike glycoprotein; 2-36 Fab heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7sbk | 3.44 Å | 2021-11-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | 42.23% | 42.48% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-09-25 | 34698504 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7sbo | 3.44 Å | 2021-11-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | 42.23% | 42.48% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-09-25 | 34698504 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7sbp | 3.44 Å | 2021-11-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | 42.23% | 42.48% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-09-25 | 34698504 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7sbq | 3.50 Å | 2021-11-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | 43.51% | 43.66% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-09-25 | 34698504 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7sbr | 3.50 Å | 2021-11-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | 43.51% | 43.66% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-09-25 | 34698504 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7sbs | 3.50 Å | 2021-11-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | 43.51% | 43.66% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-09-25 | 34698504 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7vh8 | 1.59 Å | 2021-11-03 | X-ray | H2S, 7DI | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.44% | 20.45% | 8.20% | HYDROSULFURIC ACID; PF-07321332, bound form | HYDROSULFURIC ACID; PF-07321332,bound form | XDS | 99.90% | 16.66 | SSRF (BL19U1) | PHENIX | 2021-09-21 | 34687004 | - | - | - | - | N/A | - | - | 3C-like proteinase | 54.0 | - | No | - | 78.0 | 37.5 | 67.3 | - | ||||||
| 7vmu | 2.89 Å | 2021-11-03 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 24.10% | 27.29% | 15.00% | - | HKL-3000 | 95.20% | 5.4 | SSRF (BL17U1) | PHENIX | 2021-10-09 | 35581593 | - | - | - | - | N/A | - | - | Spike protein S1; scFv E4 | 20.8 | - | No | - | 18.0 | 71.8 | 26.5 | - | |||||||
| 7ddc | 2.17 Å | 2021-11-10 | X-ray | H3F | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.12% | 23.38% | 4.70% | Tafenoquine | Tafenoquine | HKL-2000 | 99.62% | 27.382 | NSRRC (BL15A1) | REFMAC | 2020-10-28 | 35101449 | - | - | - | - | N/A | - | - | 3C-like proteinase | 56.0 | - | No | - | 51.3 | 68.5 | 66.9 | - | ||||||
| 7lfu | 2.29 Å | 2021-11-10 | X-ray | - | SARS-CoV | NSP3: PLpro | Functional ligand | 22.44% | 26.47% | 7.40% | Papain-like protease peptide inhibitor VIR250 | - | XDS | 100.00% | 16.0 | APS (24-ID-E) | PHENIX | 2021-01-18 | 34547223 | - | - | - | - | N/A | - | - | papain-like protease; Papain-like protease peptide inhibitor VIR250 | 34.9 | - | No | - | 23.3 | 64.6 | 58.1 | - | ||||||
| 7lfv | 2.23 Å | 2021-11-10 | X-ray | NH4 | SARS-CoV | NSP3: PLpro | Functional ligand | 19.72% | 23.46% | 9.90% | AMMONIUM ION; Papain-like protease peptide inhibitor VIR251 | AMMONIUM ION | XDS | 99.90% | 30.9 | APS (24-ID-E) | PHENIX | 2021-01-18 | 34547223 | ZN | - | - | - | N/A | - | - | papain-like protease; Papain-like protease peptide inhibitor VIR251 | 41.5 | - | No | - | 50.4 | 56.9 | 51.4 | - | ||||||
| 7ltn | 1.79 Å | 2021-11-10 | X-ray | YCV | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.82% | 24.49% | 12.60% | 2-[4-(1~{H}-indazol-4-yl)-2-methanoyl-6-methoxy-phenoxy]-~{N},~{N}-dimethyl-ethanamide | 2-[4-(1~{H}-indazol-4-yl)-2-methanoyl-6-methoxy-phenoxy]-~{N},~{N}-dimethyl-ethanamide | iMOSFLM | 99.43% | 6.0 | ALS (8.2.1) | PHENIX; PHENIX | 2021-02-19 | 34426525 | - | - | - | - | N/A | - | - | 3C-like proteinase | 28.4 | - | No | - | 40.4 | 35.2 | 57.0 | - | ||||||
| 7n4l | 3.60 Å | 2021-11-10 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 26.23% | 32.94% | N/A | - | HKL-2000 | 78.00% | 4.3 | APS (21-ID-E) | PHENIX | 2021-06-04 | 34716452 | - | - | - | - | N/A | - | - | WRAIR-2125 antibody Fab heavy chain; Spike protein S1 - receptor binding domain (RBD); WRAIR-2125 antibody Fab light chain | 24.5 | - | No | - | 4.4 | 95.1 | 24.9 | - | |||||||
| 7n4m | 3.79 Å | 2021-11-10 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 25.75% | 31.68% | N/A | - | HKL-2000 | 99.70% | 13.0 | APS (21-ID-E) | PHENIX | 2021-06-04 | 34716452 | - | - | - | - | N/A | - | - | WRAIR-2151 antibody Fab heavy chain; Spike protein S1 - Receptor Binding Domain (RBD); WRAIR-2151 antibody Fab light chain | 3.7 | - | No | - | 5.3 | 13.6 | 45.0 | - | |||||||
| 7opl | 4.12 Å | 2021-11-10 | Cryo-EM | SF4 | SARS-CoV | NSP1 | Protein-protein complex | N/A | N/A | N/A | IRON/SULFUR CLUSTER | IRON/SULFUR CLUSTER | - | - | () | PHENIX | 2021-06-01 | 34719824 | ZN | - | - | - | N/A | - | - | DNA primase small subunit; DNA polymerase alpha subunit B; Non-structural protein 1; DNA primase large subunit; DNA polymerase alpha catalytic subunit | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7rfr | 1.63 Å | 2021-11-10 | X-ray | 4W8 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.37% | 22.81% | N/A | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-3-(4-methoxy-1H-indole-2-carbonyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-3-(4-methoxy-1H-indole-2-carbonyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | XDS | 65.80% | 12.5 | APS (17-ID) | BUSTER | 2021-07-14 | 34726479 | - | - | - | - | N/A | - | - | 3C-like proteinase | 48.1 | - | No | - | 56.9 | 50.3 | 64.3 | - | ||||||
| 7rfs | 1.91 Å | 2021-11-10 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 21.52% | 26.04% | N/A | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | XDS | 66.10% | 10.7 | APS (17-ID) | BUSTER | 2021-07-14 | 34726479 | - | - | - | - | N/A | - | - | 3C-like proteinase | 28.9 | - | No | - | 26.2 | 35.2 | 72.2 | - | ||||||
| 7rfu | 2.50 Å | 2021-11-10 | X-ray | 4YG | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.67% | 27.61% | N/A | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-3-[N-(methanesulfonyl)-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-3-[N-(methanesulfonyl)-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | autoPROC | 66.20% | 9.7 | APS (17-ID) | BUSTER | 2021-07-14 | 34726479 | - | - | - | - | N/A | - | - | 3C-like proteinase | 14.7 | - | No | - | 16.0 | 39.2 | 46.6 | - | ||||||
| 7rfw | 1.73 Å | 2021-11-10 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.47% | 22.74% | N/A | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | autoPROC | 69.80% | 10.5 | APS (17-ID) | BUSTER | 2021-07-14 | 34726479 | - | - | - | - | N/A | - | - | 3C-like proteinase | 54.9 | - | No | - | 57.7 | 45.7 | 81.1 | - | ||||||
| 7rls | 2.00 Å | 2021-11-10 | X-ray | 5YN | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.24% | 20.10% | 7.70% | 6-[4-(3,4,5-trichlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | 6-[4-(3,4,5-trichlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | CrysalisPro | 99.90% | 14.26 | ROTATING ANODE () | PHENIX | 2021-07-26 | 34705466 | - | - | - | - | N/A | - | - | 3C-like proteinase | 45.8 | - | No | - | 81.0 | 23.0 | 63.0 | - | ||||||
| 7rm2 | 2.00 Å | 2021-11-10 | X-ray | 5YJ | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.64% | 19.01% | 8.60% | 6-[4-(3,5-dichloro-4-methylphenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | 6-[4-(3,5-dichloro-4-methylphenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | CrysalisPro | 99.90% | 15.34 | ROTATING ANODE () | PHENIX | 2021-07-26 | 34705466 | - | - | - | - | N/A | - | - | 3C-like proteinase | 68.4 | - | No | - | 87.7 | 25.3 | 99.2 | - | ||||||
| 7rmb | 2.00 Å | 2021-11-10 | X-ray | 5Z7 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 15.89% | 19.37% | 7.60% | 6-[4-(4-bromo-3-chlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | 6-[4-(4-bromo-3-chlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | CrysalisPro | 98.40% | 14.72 | ROTATING ANODE () | PHENIX | 2021-07-27 | 34705466 | - | - | - | - | N/A | - | - | 3C-like proteinase | 62.5 | - | No | - | 85.8 | 21.1 | 92.8 | - | ||||||
| 7rme | 2.00 Å | 2021-11-10 | X-ray | 5Z3 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.29% | 20.35% | 4.90% | 6-{4-[4-chloro-3-(trifluoromethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(1H,3H)-dione | 6-{4-[4-chloro-3-(trifluoromethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(1H,3H)-dione | CrysalisPro | 99.90% | 21.78 | ROTATING ANODE () | PHENIX | 2021-07-27 | 34705466 | - | - | - | - | N/A | - | - | 3C-like proteinase | 55.1 | - | No | - | 79.3 | 31.3 | 74.3 | - | ||||||
| 7rmt | 2.00 Å | 2021-11-10 | X-ray | 5ZN | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.33% | 19.57% | 6.00% | 2-chloro-4-[4-(2,6-dioxo-1,2,5,6-tetrahydropyrimidine-4-carbonyl)piperazin-1-yl]benzaldehyde | 2-chloro-4-[4-(2,6-dioxo-1,2,5,6-tetrahydropyrimidine-4-carbonyl)piperazin-1-yl]benzaldehyde | CrysalisPro | 99.90% | 17.6 | ROTATING ANODE () | PHENIX | 2021-07-28 | 34705466 | - | - | - | - | N/A | - | - | 3C-like proteinase | 64.3 | - | No | - | 84.6 | 28.0 | 90.9 | - | ||||||
| 7rmz | 2.10 Å | 2021-11-10 | X-ray | 5ZJ | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 15.22% | 18.93% | 9.00% | 6-{4-[3-chloro-4-(trifluoromethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(1H,3H)-dione | 6-{4-[3-chloro-4-(trifluoromethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(1H,3H)-dione | CrysalisPro | 99.80% | 11.01 | ROTATING ANODE () | PHENIX | 2021-07-28 | 34705466 | - | - | - | - | N/A | - | - | 3C-like proteinase | 65.0 | - | No | - | 88.1 | 23.0 | 93.8 | - | ||||||
| 7rn4 | 1.85 Å | 2021-11-10 | X-ray | H69 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.56% | 18.97% | 4.70% | 6-[4-(3,4-dichlorophenyl)piperidine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | 6-[4-(3,4-dichlorophenyl)piperidine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | CrysalisPro | 98.20% | 24.07 | ROTATING ANODE () | PHENIX | 2021-07-29 | 34705466 | - | - | - | - | N/A | - | - | 3C-like proteinase | 53.5 | - | No | - | 88.0 | 21.1 | 72.7 | - | ||||||
| 7rnh | 2.00 Å | 2021-11-10 | X-ray | 5ZW | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.14% | 20.05% | 5.90% | 6-[4-(4-chlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | 6-[4-(4-chlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | CrysalisPro | 99.60% | 21.2 | ROTATING ANODE () | PHENIX | 2021-07-29 | 34705466 | - | - | - | - | N/A | - | - | 3C-like proteinase | 55.3 | - | No | - | 81.4 | 23.0 | 80.9 | - | ||||||
| 7rnk | 2.10 Å | 2021-11-10 | X-ray | 5ZT | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.56% | 20.64% | 9.90% | 6-{4-[3-chloro-4-(hydroxymethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(3H,5H)-dione | 6-{4-[3-chloro-4-(hydroxymethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(3H,5H)-dione | CrysalisPro | 99.90% | 11.47 | ROTATING ANODE () | PHENIX | 2021-07-29 | 34705466 | - | - | - | - | N/A | - | - | 3C-like proteinase | 54.0 | - | No | - | 77.0 | 16.8 | 88.9 | - | ||||||
| 7sbl | 3.44 Å | 2021-11-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | 42.23% | 42.48% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-09-25 | 34698504 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7sbt | 3.50 Å | 2021-11-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | 43.51% | 43.66% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-09-25 | 34698504 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7sg4 | 3.43 Å | 2021-11-10 | Cryo-EM | NAG | SARS-CoV | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-10-04 | 34726473 | - | - | - | - | N/A | - | - | Spike glycoprotein; DH1047 Heavy chain; DH1047 light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7bnv | 2.35 Å | 2021-11-17 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 22.36% | 26.46% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.08% | 16.34 | ESRF (MASSIF-1) | REFMAC; PHENIX | 2021-01-22 | 34211469 | - | - | - | - | N/A | - | - | Light Chain; Heavy Chain; Surface glycoprotein | 15.8 | - | No | - | 23.3 | 14.8 | 66.6 | - | ||||||
| 7dfg | 2.70 Å | 2021-11-17 | Cryo-EM | 1RP, POP | SARS-CoV-2 | NSP7/NSP8/NSP12 | Functional ligand | N/A | N/A | N/A | 6-fluoro-3-oxo-4-(5-O-phosphono-beta-D-ribofuranosyl)-3,4-dihydropyrazine-2-carboxamide; RNA (5'-R(P*AP*GP*AP*UP*UP*AP*AP*GP*UP*UP*AP*U)-3'); RNA (5'-R(P*CP*CP*CP*UP*AP*UP*AP*AP*CP*UP*UP*AP*AP*UP*CP*U)-3'); PYROPHOSPHATE 2- | 6-fluoro-3-oxo-4-(5-O-phosphono-beta-D-ribofuranosyl)-3,4-dihydropyrazine-2-carboxamide; PYROPHOSPHATE 2- | - | - | () | PHENIX | 2020-11-08 | - | ZN; MG | - | - | - | N/A | - | - | RNA (5'-R(P*AP*GP*AP*UP*UP*AP*AP*GP*UP*UP*AP*U)-3'); RNA (5'-R(P*CP*CP*CP*UP*AP*UP*AP*AP*CP*UP*UP*AP*AP*UP*CP*U)-3'); RNA-directed RNA polymerase; Non-structural protein 8; Non-structural protein 7 | N/A | - | No | - | - | N/A | N/A | N/A | RNA (5'-R(P*AP*GP*AP*UP*UP*AP*AP*GP*UP*UP*AP*U)-3'); RNA (5'-R(P*CP*CP*CP*UP*AP*UP*AP*AP*CP*UP*UP*AP*AP*UP*CP*U)-3') | - | |||||
| 7dfh | 2.97 Å | 2021-11-17 | Cryo-EM | POP, RVP | SARS-CoV-2 | NSP7/NSP8/NSP12 | Functional ligand | N/A | N/A | N/A | PYROPHOSPHATE 2-; RNA (5'-R(P*CP*CP*CP*CP*CP*AP*CP*AP*UP*AP*GP*C)-3'); RNA (5'-R(P*GP*CP*UP*AP*UP*GP*UP*G*(LIG))-3'); RIBAVIRIN MONOPHOSPHATE | PYROPHOSPHATE 2-; RIBAVIRIN MONOPHOSPHATE | - | - | () | 2020-11-08 | - | ZN; MG | - | - | - | N/A | - | - | RNA (5'-R(P*CP*CP*CP*CP*CP*AP*CP*AP*UP*AP*GP*C)-3'); RNA-directed RNA polymeras; Non-structural protein 8; Non-structural protein 7; RNA (5'-R(P*GP*CP*UP*AP*UP*GP*UP*G*(LIG))-3') | N/A | - | No | - | - | N/A | N/A | N/A | RNA (5'-R(P*GP*CP*UP*AP*UP*GP*UP*G*(LIG))-3'); RNA (5'-R(P*CP*CP*CP*CP*CP*AP*CP*AP*UP*AP*GP*C)-3') | - | ||||||
| 7e5s | 3.60 Å | 2021-11-17 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-02-20 | 33782529 | - | - | - | - | N/A | - | - | P17 light chain; FC05 light chain; FC05 heavy chain; HB27 heavy chain; HB27 light chain; Spike glycoprotein; H014 light chain; H014 heavy chain; P17 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7e9t | 10.90 Å | 2021-11-17 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-03-05 | 34782481 | - | - | - | - | N/A | - | - | Spike protein S2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7np1 | 2.80 Å | 2021-11-17 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 23.21% | 28.10% | 6.29% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.75% | 5.79 | ESRF (MASSIF-1) | PDB-REDO; PHENIX | 2021-02-26 | 34211469 | - | - | - | - | N/A | - | - | Immunoblobulin light chain; Immunoglobulin gamma-1 heavy chain; Spike protein S1 | 10.7 | - | No | - | 13.5 | 42.2 | 34.7 | - | ||||||
| 7so9 | 2.40 Å | 2021-11-17 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-10-29 | 34751595 | - | - | - | - | N/A | - | - | Spike glycoprotein; S2M11 Fab heavy chain; S2L20 Fab light chain; S2L20 Fab heavy chain; S2M11 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7soa | 3.10 Å | 2021-11-17 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-10-29 | 34751595 | - | - | - | - | N/A | - | - | S2L20 Fab heavy chain; Spike glycoprotein; S2L20 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7sob | 2.40 Å | 2021-11-17 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-10-29 | 34751595 | - | - | - | - | N/A | - | - | S2L20 Fab heavy chain; Spike glycoprotein; S309 Fab heavy chain; S2L20 Fab light chain; S309 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7soc | 3.30 Å | 2021-11-17 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-10-29 | 34751595 | - | - | - | - | N/A | - | - | S309 Fab light chain; S309 Fab heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7sod | 3.20 Å | 2021-11-17 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-10-29 | 34751595 | - | - | - | - | N/A | - | - | S2L20 Fab heavy chain; Spike glycoprotein; S2L20 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7soe | 2.80 Å | 2021-11-17 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-10-29 | 34751595 | - | - | - | - | N/A | - | - | Spike glycoprotein; S2X303 Fab heavy chain; S2X303 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7sof | 3.60 Å | 2021-11-17 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-10-29 | 34751595 | - | - | - | - | N/A | - | - | Spike glycoprotein; S2X303 Fab heavy chain; S2X303 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7sqe | 2.00 Å | 2021-11-17 | X-ray | JWX | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 17.39% | 20.47% | 9.10% | (1R)-N-[(1H-indol-3-yl)methyl]-N-methyl-1-(naphthalen-1-yl)ethan-1-amine | (1R)-N-[(1H-indol-3-yl)methyl]-N-methyl-1-(naphthalen-1-yl)ethan-1-amine | HKL-3000 | 98.40% | 8.0 | APS (19-ID) | REFMAC | 2021-11-05 | - | ZN | - | - | - | N/A | - | - | Papain-like protease | 74.6 | - | No | - | - | 78.4 | 55.7 | 92.7 | - | |||||
| 6x45 | 2.20 Å | 2021-11-24 | X-ray | - | SARS-CoV-2 | Spike | Functional ligand | 24.29% | 27.84% | N/A | Spike protein S2'; Spike protein S2' | - | XDS | 99.50% | 9.0 | NSLS-II (17-ID-1) | PHENIX | 2020-05-22 | - | - | - | - | - | N/A | - | - | Spike protein S2'; Spike protein S2' | 20.5 | - | No | - | - | 14.9 | 12.8 | 87.9 | - | |||||
| 7dgb | 1.68 Å | 2021-11-24 | X-ray | EOF | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.35% | 23.89% | 11.80% | (2~{S})-4-methyl-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pentanamide | (2~{S})-4-methyl-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pentanamide | HKL-3000 | 98.10% | 17.11 | SSRF (BL19U1) | PHENIX | 2020-11-11 | 35635946 | - | - | - | - | N/A | - | - | 3C-like proteinase | 32.5 | - | No | - | 46.1 | 26.3 | 68.7 | - | ||||||
| 7dgf | 1.64 Å | 2021-11-24 | X-ray | H60 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.98% | 21.82% | 4.20% | (2~{S})-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepiperidin-3-yl]propan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]hexanamide | (2~{S})-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepiperidin-3-yl]propan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]hexanamide | HKL-3000 | 96.60% | 41.33 | ROTATING ANODE () | PHENIX | 2020-11-11 | 35635946 | - | - | - | - | N/A | - | - | 3C-like proteinase | 38.9 | - | No | - | 66.8 | 25.0 | 62.0 | - | ||||||
| 7dgg | 2.00 Å | 2021-11-24 | X-ray | H63 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.61% | 21.69% | 4.70% | (2~{S})-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]hexanamide | (2~{S})-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]hexanamide | HKL-3000 | 92.50% | 34.28 | SSRF (BL19U1) | PHENIX | 2020-11-11 | 35635946 | - | - | - | - | N/A | - | - | 3C-like proteinase | 63.1 | - | No | - | 68.2 | 71.0 | 61.7 | - | ||||||
| 7dgh | 1.97 Å | 2021-11-24 | X-ray | H6F | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.58% | 23.52% | 11.50% | ~{N}-[(2~{S})-3-methyl-1-[[(2~{S})-4-methyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepiperidin-3-yl]propan-2-yl]amino]pentan-2-yl]amino]-1-oxidanylidene-butan-2-yl]naphthalene-2-carboxamide | ~{N}-[(2~{S})-3-methyl-1-[[(2~{S})-4-methyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepiperidin-3-yl]propan-2-yl]amino]pentan-2-yl]amino]-1-oxidanylidene-butan-2-yl]naphthalene-2-carboxamide | HKL-3000 | 98.60% | 11.4 | SSRF (BL19U1) | PHENIX | 2020-11-11 | 35635946 | - | - | - | - | N/A | - | - | 3C-like proteinase | 21.1 | - | No | - | 49.7 | 22.0 | 45.3 | - | ||||||
| 7dgi | 1.90 Å | 2021-11-24 | X-ray | H6L | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.27% | 21.29% | 17.90% | ~{N}-[(2~{S})-3-methyl-1-[[(2~{S})-4-methyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepiperidin-3-yl]propan-2-yl]amino]pentan-2-yl]amino]-1-oxidanylidene-butan-2-yl]-4-nitro-benzamide | ~{N}-[(2~{S})-3-methyl-1-[[(2~{S})-4-methyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepiperidin-3-yl]propan-2-yl]amino]pentan-2-yl]amino]-1-oxidanylidene-butan-2-yl]-4-nitro-benzamide | HKL-3000 | 100.00% | 9.52 | SSRF (BL19U1) | PHENIX | 2020-11-11 | 35635946 | - | - | - | - | N/A | - | - | 3C-like proteinase | 50.9 | - | No | - | 71.7 | 42.8 | 62.4 | - | ||||||
| 7dhj | 1.96 Å | 2021-11-24 | X-ray | H6R | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.90% | 23.09% | 7.90% | (2~{S})-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pent-4-ynamide | (2~{S})-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pent-4-ynamide | HKL-3000 | 96.70% | 21.43 | SSRF (BL19U1) | PHENIX | 2020-11-15 | 35635946 | - | - | - | - | N/A | - | - | 3C-like proteinase | 37.8 | - | No | - | 54.3 | 29.2 | 68.1 | - | ||||||
| 7efp | 2.70 Å | 2021-11-24 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 18.38% | 22.42% | 15.50% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 100.00% | 19.6 | SSRF (BL18U1) | PHENIX | 2021-03-22 | 34531369 | - | - | - | - | N/A | - | - | Spike glycoprotein - UNP 320-537; Processed angiotensin-converting enzyme 2 | 46.6 | - | No | - | 61.0 | 75.5 | 32.1 | - | ||||||
| 7efr | 2.49 Å | 2021-11-24 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 19.01% | 22.89% | 12.60% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 100.00% | 26.0 | SSRF (BL18U1) | PHENIX | 2021-03-23 | 34531369 | - | - | - | - | N/A | - | - | Spike glycoprotein - UNP residues 321-537; Processed angiotensin-converting enzyme 2 | 36.6 | - | No | - | 56.3 | 52.1 | 41.0 | - | ||||||
| 7rdx | 3.10 Å | 2021-11-24 | Cryo-EM | 1N7, ADP, AF3 | SARS-CoV-2 | NSP7/NSP8/NSP12/NSP13 | Protein-protein complex | N/A | N/A | N/A | ADENOSINE-5'-DIPHOSPHATE; CHAPSO; ALUMINUM FLUORIDE; Product RNA; Template RNA | CHAPSO; ADENOSINE-5'-DIPHOSPHATE; ALUMINUM FLUORIDE | - | - | () | PHENIX | 2021-07-12 | 35260847 | ZN; MG | - | - | - | N/A | - | - | Non-structural protein 7 - UNP residues 3860-3942; RNA-directed RNA polymerase - UNP residues 4393-5324; Non-structural protein 8 - UNP residues 3943-4140; Helicase - UNP residues 5325-5925; Product RNA; Template RNA | N/A | - | No | - | N/A | N/A | N/A | Product RNA; Template RNA | - | ||||||
| 7rdz | 3.60 Å | 2021-11-24 | Cryo-EM | ADP | SARS-CoV-2 | NSP7/NSP8/NSP12/NSP13 | Protein-protein complex | N/A | N/A | N/A | Template RNA; ADENOSINE-5'-DIPHOSPHATE; Product RNA | ADENOSINE-5'-DIPHOSPHATE | - | - | () | PHENIX | 2021-07-12 | 35260847 | ZN; MG | - | - | - | N/A | - | - | RNA-directed RNA polymerase - UNP residues 4393-5324; Template RNA; Product RNA; Helicase - UNP residues 5325-5925; Non-structural protein 7 - UNP residues 3860-3942; Non-structural protein 8 - UNP residues 3943-4140 | N/A | - | No | - | N/A | N/A | N/A | Product RNA; Template RNA | - | ||||||
| 7re3 | 3.33 Å | 2021-11-24 | Cryo-EM | ADP, 1N7, AF3 | SARS-CoV-2 | NSP7/NSP8/NSP12/NSP13 | Protein-protein complex | N/A | N/A | N/A | ADENOSINE-5'-DIPHOSPHATE; Template RNA; Product RNA; CHAPSO; ALUMINUM FLUORIDE | ADENOSINE-5'-DIPHOSPHATE; CHAPSO; ALUMINUM FLUORIDE | - | - | () | PHENIX | 2021-07-12 | 35260847 | ZN; MG | - | - | - | N/A | - | - | Product RNA; RNA-directed RNA polymerase - UNP residues 4393-5324; Non-structural protein 8 - UNP residues 3943-4140; Non-structural protein 7 - UNP residues 3860-3942; Helicase - UNP residues 5325-5925; Template RNA | N/A | - | No | - | N/A | N/A | N/A | Product RNA; Template RNA | - | ||||||
| 7vnb | 2.27 Å | 2021-11-24 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 17.67% | 20.33% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.90% | 11.1 | SSRF (BL17U1) | PHENIX | 2021-10-10 | 34732694 | - | - | - | - | N/A | - | - | Spike protein S1; n3113 | 66.3 | - | No | - | 79.4 | 66.7 | 61.6 | - | ||||||
| 7vnc | 3.70 Å | 2021-11-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-10-10 | 34732694 | - | - | - | - | N/A | - | - | Spike glycoprotein; n3113 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7vnd | 3.60 Å | 2021-11-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-10-10 | 34732694 | - | - | - | - | N/A | - | - | n3113; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7vne | 3.50 Å | 2021-11-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-10-10 | 34732694 | - | - | - | - | N/A | - | - | n3113.1; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7vx1 | 3.50 Å | 2021-11-24 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-11-12 | 34930910 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7vx9 | 4.00 Å | 2021-11-24 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-11-12 | 34930910 | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7vxa | 3.90 Å | 2021-11-24 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-11-12 | 34930910 | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7b0b | 2.98 Å | 2021-12-01 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 20.90% | 24.62% | N/A | - | HKL-3000 | 77.27% | 4.1 | ROTATING ANODE () | PHENIX | 2020-11-19 | - | - | - | - | - | N/A | - | - | Surface glycoprotein; IGK@ protein; Fab heavy chain | 30.3 | - | No | - | - | 38.9 | 81.7 | 16.0 | - | ||||||
| 7dg6 | 2.40 Å | 2021-12-01 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 17.54% | 23.26% | N/A | - | XDS | 99.47% | 10.54 | ROTATING ANODE () | PHENIX | 2020-11-11 | - | - | - | - | - | N/A | - | - | Replicase polyprotein 1ab - UNP residues 3264-3565 | 58.1 | - | No | - | - | 52.5 | 48.5 | 89.8 | - | ||||||
| 7rdy | 3.10 Å | 2021-12-01 | Cryo-EM | ADP, AF3, 1N7 | SARS-CoV-2 | NSP7/NSP8/NSP12/NSP13 | Protein-protein complex | N/A | N/A | N/A | Template RNA; CHAPSO; ALUMINUM FLUORIDE; Product RNA; ADENOSINE-5'-DIPHOSPHATE | ADENOSINE-5'-DIPHOSPHATE; ALUMINUM FLUORIDE; CHAPSO | - | - | () | PHENIX | 2021-07-12 | 35260847 | ZN; MG | - | - | - | N/A | - | - | Template RNA; RNA-directed RNA polymerase - UNP residues 4393-5324; Non-structural protein 8 - UNP residues 3943-4140; Non-structural protein 7 - UNP residues 3860-3942; Helicase; Product RNA | N/A | - | No | - | N/A | N/A | N/A | Product RNA; Template RNA | - | ||||||
| 7re0 | 3.50 Å | 2021-12-01 | Cryo-EM | AF3, ADP | SARS-CoV-2 | NSP7/NSP8/NSP12/NSP13 | Protein-protein complex | N/A | N/A | N/A | ALUMINUM FLUORIDE; ADENOSINE-5'-DIPHOSPHATE; Template RNA; Product RNA | ALUMINUM FLUORIDE; ADENOSINE-5'-DIPHOSPHATE | - | - | () | PHENIX | 2021-07-12 | 35260847 | MG; ZN | - | - | - | N/A | - | - | Product RNA; RNA-directed RNA polymerase - UNP residues 4393-5324; Non-structural protein 8 - UNP residues 3943-4140; Non-structural protein 7 - UNP residues 3860-3942; Helicase - UNP residues 5325-5925; Template RNA | N/A | - | No | - | N/A | N/A | N/A | Template RNA; Product RNA | - | ||||||
| 7re1 | 2.91 Å | 2021-12-01 | Cryo-EM | AF3, 1N7, ADP | SARS-CoV-2 | NSP7/NSP8/NSP12/NSP13 | Protein-protein complex | N/A | N/A | N/A | ADENOSINE-5'-DIPHOSPHATE; Template RNA; Product RNA; ALUMINUM FLUORIDE; CHAPSO | ALUMINUM FLUORIDE; CHAPSO; ADENOSINE-5'-DIPHOSPHATE | - | - | () | PHENIX | 2021-07-12 | 35260847 | MG; ZN | - | - | - | N/A | - | - | Product RNA; RNA-directed RNA polymerase - UNP residues 4393-5324; Non-structural protein 8 - UNP residues 3943-4140; Non-structural protein 7 - UNP residues 3860-3942; Helicase; Template RNA | N/A | - | No | - | N/A | N/A | N/A | Template RNA; Product RNA | - | ||||||
| 7re2 | 3.17 Å | 2021-12-01 | Cryo-EM | 1N7, ADP, AF3 | SARS-CoV-2 | NSP7/NSP8/NSP12/NSP13 | Protein-protein complex | N/A | N/A | N/A | ALUMINUM FLUORIDE; CHAPSO; Template RNA; Product RNA; ADENOSINE-5'-DIPHOSPHATE | CHAPSO; ADENOSINE-5'-DIPHOSPHATE; ALUMINUM FLUORIDE | - | - | () | PHENIX | 2021-07-12 | 35260847 | ZN; MG | - | - | - | N/A | - | - | Helicase - UNP residues 5325-5925; RNA-directed RNA polymerase - UNP residues 4393-5324; Non-structural protein 8 - UNP residues 3943-4140; Template RNA; Product RNA; Non-structural protein 7 - UNP residues 3860-3942 | N/A | - | No | - | N/A | N/A | N/A | Product RNA; Template RNA | - | ||||||
| 7vx4 | 3.90 Å | 2021-12-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-12 | 34930910 | ZN | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7vx5 | 3.80 Å | 2021-12-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-12 | 34930910 | ZN | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7vxd | 4.00 Å | 2021-12-01 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-11-12 | 34930910 | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7vxe | 3.20 Å | 2021-12-01 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-11-12 | 34930910 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7vxf | 3.60 Å | 2021-12-01 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-11-12 | 34930910 | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7dk2 | 3.00 Å | 2021-12-08 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 25.77% | 30.39% | 19.90% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 98.50% | 4.0 | SSRF (BL19U1) | PHENIX | 2020-11-22 | - | - | - | - | - | N/A | - | - | MW07 heavy chain; MW07 light chain; Spike protein S1 - Receptor Binding Domain | 28.0 | - | No | - | - | 6.7 | 74.8 | 50.1 | - | |||||
| 7sn0 | 3.08 Å | 2021-12-08 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 23.47% | 28.60% | 24.60% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 98.90% | 5.29 | APS (24-ID-C) | BUSTER | 2021-10-27 | 34855508 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Surface glycoprotein | 22.1 | - | No | - | 11.4 | 75.5 | 32.3 | - | ||||||
| 7sn2 | 4.30 Å | 2021-12-08 | Cryo-EM | - | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-10-27 | 34855508 | - | - | - | - | N/A | - | - | neutralizing antibody C1C-A3 Fab light chain; neutralizing antibody C1C-A3 Fab heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7sn3 | 3.10 Å | 2021-12-08 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-10-27 | 34855508 | - | - | - | - | N/A | - | - | neutralizing antibody C1C-A3 heavy chain variable region; neutralizing antibody C1C-A3 light chain variable region; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7doi | 2.60 Å | 2021-12-15 | Cryo-EM | POP, HCU | SARS-CoV-2 | NSP7/NSP8/NSP12 | Functional ligand | N/A | N/A | N/A | RNA (5'-R(P*AP*GP*AP*UP*UP*AP*AP*GP*UP*UP*AP*U)-3'); [(2R)-4-(2-azanyl-6-oxidanylidene-3H-purin-9-yl)-2-(hydroxymethyl)butyl] dihydrogen phosphate; PYROPHOSPHATE 2-; RNA (5'-R(P*CP*CP*UP*AP*UP*AP*AP*CP*UP*UP*AP*AP*UP*CP*U)-3') | PYROPHOSPHATE 2-; [(2R)-4-(2-azanyl-6-oxidanylidene-3H-purin-9-yl)-2-(hydroxymethyl)butyl] dihydrogen phosphate | - | - | () | PHENIX | 2020-12-14 | - | ZN; MG | - | - | - | N/A | - | - | RNA (5'-R(P*AP*GP*AP*UP*UP*AP*AP*GP*UP*UP*AP*U)-3'); RNA (5'-R(P*CP*CP*UP*AP*UP*AP*AP*CP*UP*UP*AP*AP*UP*CP*U)-3'); Non-structural protein 7; Non-structural protein 8; RNA-directed RNA polymerase | N/A | - | No | - | - | N/A | N/A | N/A | RNA (5'-R(P*AP*GP*AP*UP*UP*AP*AP*GP*UP*UP*AP*U)-3'); RNA (5'-R(P*CP*CP*UP*AP*UP*AP*AP*CP*UP*UP*AP*AP*UP*CP*U)-3') | - | |||||
| 7dok | 2.73 Å | 2021-12-15 | Cryo-EM | HCU, POP | SARS-CoV-2 | NSP7/NSP8/NSP12 | Functional ligand | N/A | N/A | N/A | [(2R)-4-(2-azanyl-6-oxidanylidene-3H-purin-9-yl)-2-(hydroxymethyl)butyl] dihydrogen phosphate; RNA (5'-R(P*CP*CP*CP*UP*AP*UP*AP*AP*CP*UP*UP*AP*AP*UP*CP*UP*CP*AP*CP*AP*UP*AP*GP*C)-3'); RNA (5'-R(P*GP*CP*UP*AP*UP*GP*UP*GP*AP*GP*AP*UP*UP*AP*AP*GP*UP*UP*AP*U)-3'); PYROPHOSPHATE 2- | [(2R)-4-(2-azanyl-6-oxidanylidene-3H-purin-9-yl)-2-(hydroxymethyl)butyl] dihydrogen phosphate; PYROPHOSPHATE 2- | - | - | () | PHENIX | 2020-12-14 | - | MG; ZN | - | - | - | N/A | - | - | RNA (5'-R(P*GP*CP*UP*AP*UP*GP*UP*GP*AP*GP*AP*UP*UP*AP*AP*GP*UP*UP*AP*U)-3'); Non-structural protein 7; Non-structural protein 8; RNA-directed RNA polymerase; RNA (5'-R(P*CP*CP*CP*UP*AP*UP*AP*AP*CP*UP*UP*AP*AP*UP*CP*UP*CP*AP*CP*AP*UP*AP*GP*C)-3') | N/A | - | No | - | - | N/A | N/A | N/A | RNA (5'-R(P*GP*CP*UP*AP*UP*GP*UP*GP*AP*GP*AP*UP*UP*AP*AP*GP*UP*UP*AP*U)-3'); RNA (5'-R(P*CP*CP*CP*UP*AP*UP*AP*AP*CP*UP*UP*AP*AP*UP*CP*UP*CP*AP*CP*AP*UP*AP*GP*C)-3') | - | |||||
| 7fem | 4.10 Å | 2021-12-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-07-21 | 34750362 | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7fet | 3.70 Å | 2021-12-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-07-21 | 34750362 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7pry | 3.10 Å | 2021-12-15 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 24.45% | 29.09% | 24.60% | - | xia2 | 100.00% | 6.6 | Diamond (I03) | PHENIX | 2021-09-22 | 34921776 | - | - | - | - | N/A | - | - | COVOX-45 Fab light chain; COVOX-45 Fab heavy chain; Spike protein S1; Beta-6 Fab light chain; Beta-6 Fab heavy chain | 14.4 | - | No | - | 9.7 | 33.6 | 57.9 | - | |||||||
| 7prz | 3.20 Å | 2021-12-15 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 19.04% | 22.29% | 47.30% | - | xia2 | 99.80% | 9.4 | Diamond (I03) | PHENIX | 2021-09-22 | 34921776 | - | - | - | - | N/A | - | - | Beta-22 Fab light chain; Beta-22 Fab heavy chain; Spike protein S1 | 32.3 | - | No | - | 62.4 | 19.1 | 59.0 | - | |||||||
| 7ps0 | 2.92 Å | 2021-12-15 | X-ray | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 22.21% | 27.05% | 48.20% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 99.70% | 4.8 | Diamond (I03) | PHENIX | 2021-09-22 | 34921776 | - | - | - | - | N/A | - | - | Spike protein S1; Beta-24 light chain; Beta-24 heavy chain | 27.7 | - | No | - | 19.2 | 47.1 | 64.7 | - | ||||||
| 7ps1 | 2.40 Å | 2021-12-15 | X-ray | IOD, NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | 18.39% | 21.51% | 18.10% | IODIDE ION; 2-acetamido-2-deoxy-beta-D-glucopyranose | IODIDE ION; 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 96.70% | 8.6 | Diamond (I03) | PHENIX | 2021-09-22 | 34921776 | - | - | - | - | N/A | - | - | Spike protein S1; Beta-27 Fab light chain; Beta-27 Fab heavy chain | 64.6 | - | No | - | 69.6 | 43.3 | 91.0 | - | ||||||
| 7ps2 | 2.99 Å | 2021-12-15 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 23.31% | 26.61% | 29.30% | - | xia2 | 99.90% | 8.5 | Diamond (I03) | PHENIX | 2021-09-22 | 34921776 | - | - | - | - | N/A | - | - | Beta-29 Fab light chain; Beta-53 Fab light chain; Beta-53 heavy chain; Beta-29 Fab heavy chain; Spike protein S1 | 7.7 | - | No | - | 22.1 | 12.1 | 46.6 | - | |||||||
| 7ps4 | 1.94 Å | 2021-12-15 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 20.35% | 23.12% | 6.50% | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | - | xia2 | 98.00% | 17.1 | Diamond (I03) | PHENIX | 2021-09-22 | 34921776 | - | - | - | - | N/A | - | - | Beta-38 Fab heavy chain; Beta-38 Fab light chain; Spike protein S1 | - | - | No | - | 53.8 | - | 80.5 | - | ||||||
| 7ps5 | 3.14 Å | 2021-12-15 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 22.55% | 27.38% | 22.90% | - | xia2 | 100.00% | 7.8 | Diamond (I03) | PHENIX | 2021-09-22 | 34921776 | - | - | - | - | N/A | - | - | Beta-47 Fab light chain; Spike protein S1; Beta-47 Fab heavy chain | 30.3 | - | No | - | 17.4 | 55.3 | 63.8 | - | |||||||
| 7ps6 | 2.26 Å | 2021-12-15 | X-ray | PG0 | SARS-CoV-2 | Spike | Pathogen-host interaction | 21.53% | 24.01% | 9.60% | 2-(2-METHOXYETHOXY)ETHANOL | 2-(2-METHOXYETHOXY)ETHANOL | xia2 | 99.50% | 14.2 | Diamond (I03) | PHENIX | 2021-09-22 | 34921776 | - | - | - | - | N/A | - | - | Spike protein S1; Beta-44 Fab light chain; Beta-44 Fab heavy chain; Beta-54 Fab heavy chain; Beta-54 Fab light chain | 57.5 | - | No | - | 44.7 | 63.4 | 81.5 | - | ||||||
| 7ps7 | 3.90 Å | 2021-12-15 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 23.33% | 28.40% | 38.90% | - | xia2 | 99.40% | 4.9 | Diamond (I03) | PHENIX | 2021-09-22 | 34921776 | - | - | - | - | N/A | - | - | Beta-40 Fab light chain; Beta-40 heavy chain; Spike protein S1 | 5.7 | - | No | - | 12.3 | 29.8 | 31.0 | - | |||||||
| 7q6e | 2.70 Å | 2021-12-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-11-07 | 34921776 | - | - | - | - | N/A | - | - | Spike glycoprotein; Beta-49 light chain; Beta-49 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7q9f | 3.60 Å | 2021-12-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-11-12 | 34921776 | - | - | - | - | N/A | - | - | Spike glycoprotein; Beta-50 light chain; Beta-50 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7q9g | 3.30 Å | 2021-12-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-11-12 | 34921776 | - | - | - | - | N/A | - | - | Spike glycoprotein; COVOX-222 heavy chain; COVOX-222 light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7q9i | 4.90 Å | 2021-12-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-11-12 | 34921776 | - | - | - | - | N/A | - | - | Spike glycoprotein; Beta-43 heavy chain; Beta-43 light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7q9j | 4.00 Å | 2021-12-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-11-12 | 34921776 | - | - | - | - | N/A | - | - | Spike glycoprotein; Beta-26 heavy chain; Beta-26 light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7q9k | 4.50 Å | 2021-12-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-11-12 | 34921776 | - | - | - | - | N/A | - | - | Beta-32 light chain; Spike glycoprotein; Beta-32 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7q9m | 3.70 Å | 2021-12-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-12 | 34921776 | - | - | - | - | N/A | - | - | Spike glycoprotein; Beta-53 fab heavy chain; Beta-53 fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7q9p | 4.50 Å | 2021-12-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-11-12 | 34921776 | - | - | - | - | N/A | - | - | Spike glycoprotein; Beta-06 light chain; Beta-06 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7qg7 | 1.72 Å | 2021-12-15 | X-ray | U08 | SARS-CoV-2 | NSP3: Macro | Functional ligand | 18.31% | 22.58% | N/A | (2~{R},3~{R},4~{S},5~{R})-2-(4-azanylpyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolane-2-carbonitrile | (2~{R},3~{R},4~{S},5~{R})-2-(4-azanylpyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolane-2-carbonitrile | XDS | 99.90% | 12.09 | BESSY (14.2) | PHENIX; PHENIX | 2021-12-07 | 35839840 | - | - | - | - | N/A | - | - | ORF1a polyprotein | 63.1 | - | No | - | 59.5 | 63.4 | 78.0 | - | ||||||
| 7rpv | 3.54 Å | 2021-12-15 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 24.35% | 29.18% | 22.00% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-3000 | 97.70% | 3.5 | SSRL (BL9-2) | PHENIX | 2021-08-04 | 34845451 | ZN | - | - | - | N/A | - | - | Spike protein S1; Processed angiotensin-converting enzyme 2 | 34.7 | - | No | - | 9.4 | 93.4 | 42.7 | - | ||||||
| 7fae | 3.65 Å | 2021-12-22 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-07-06 | 34966387 | - | - | - | - | N/A | - | - | P36-5D2 heavy chain; Spike glycoprotein; P36-5D2 light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7faf | 3.69 Å | 2021-12-22 | Cryo-EM | - | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | - | - | - | () | 2021-07-06 | 34966387 | - | - | - | - | N/A | - | - | P36-5D2 light chain; Spike glycoprotein; P36-5D2 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7mlf | 2.60 Å | 2021-12-22 | X-ray | C7A | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 22.04% | 27.85% | N/A | N-(4-tert-butylphenyl)-2-chloro-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]acetamide | N-(4-tert-butylphenyl)-2-chloro-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]acetamide | DIALS | 93.18% | 6.0 | CLSI (08B1-1) | REFMAC | 2021-04-28 | 34995923 | - | - | - | - | N/A | - | - | 3C-like proteinase - UNP residues 3264-3567 | 57.0 | - | No | - | 14.8 | 78.5 | 95.3 | - | ||||||
| 7mlg | 2.50 Å | 2021-12-22 | X-ray | ZJ1 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 22.40% | 26.80% | N/A | (2R)-2-[(4-tert-butylphenyl)(ethanesulfonyl)amino]-N-cyclohexyl-2-(pyridin-3-yl)acetamide | (2R)-2-[(4-tert-butylphenyl)(ethanesulfonyl)amino]-N-cyclohexyl-2-(pyridin-3-yl)acetamide | DIALS | 97.90% | 12.52 | APS (24-ID-C) | REFMAC | 2021-04-28 | 34995923 | - | - | - | - | N/A | - | - | 3C-like proteinase - UNP residues 3264-3569 | 34.1 | - | No | - | 20.8 | 45.4 | 78.1 | - | ||||||
| 7q0g | 1.82 Å | 2021-12-22 | X-ray | K, TAR | SARS-CoV-2 | Spike | Pathogen-host interaction | 19.17% | 22.41% | 9.50% | POTASSIUM ION; D(-)-TARTARIC ACID | POTASSIUM ION; D(-)-TARTARIC ACID | xia2 | 94.90% | 18.3 | Diamond (I03) | PHENIX | 2021-10-14 | 34921776 | K | - | - | - | N/A | - | - | FI-3A Fab heavy chain; Beta-49 Fab light chain; Beta-49 Fab heavy chain; Spike protein S1; FI-3A Fab light chain | 67.1 | - | No | - | 61.0 | 66.2 | 82.1 | - | ||||||
| 7q0h | 3.65 Å | 2021-12-22 | X-ray | - | SARS-CoV-2 | Spike | Pathogen-host interaction | 25.47% | 30.28% | 55.00% | - | xia2 | 100.00% | 3.7 | Diamond (I03) | PHENIX | 2021-10-14 | 34921776 | - | - | - | - | N/A | - | - | Beta-50 Fab light chain; Beta-54 Fab light chain; Beta-54 Fab heavy chain; Spike protein S1; Beta-50 Fab heavy chain | 12.1 | - | No | - | 6.9 | 28.8 | 59.2 | - | |||||||
| 7q0i | 2.39 Å | 2021-12-22 | X-ray | PO4, NAG, PG4 | SARS-CoV-2 | Spike | Pathogen-host interaction | 20.19% | 24.20% | 14.40% | 2-acetamido-2-deoxy-beta-D-glucopyranose; PHOSPHATE ION | PHOSPHATE ION; 2-acetamido-2-deoxy-beta-D-glucopyranose; TETRAETHYLENE GLYCOL | xia2 | 93.60% | 9.8 | Diamond (I03) | PHENIX | 2021-10-14 | 34921776 | - | - | - | - | N/A | - | - | Spike protein S1; Beta-43 Fab light chain; Beta-43 heavy chain | 27.8 | - | No | - | 42.9 | 20.0 | 68.3 | - | ||||||
| 7t3y | 1.90 Å | 2021-12-22 | X-ray | F8C, F5L | MERS-CoV | NSP5 (3CLpro) | Functional ligand | 20.50% | 25.80% | 6.00% | (1R,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; (1R,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | XDS | 99.90% | 14.8 | APS (17-ID) | PHENIX | 2021-12-09 | 35638577 | - | - | - | - | N/A | - | - | 3C-like proteinase - Full Length | 21.2 | - | No | - | 28.1 | 27.7 | 61.3 | - | ||||||
| 7t3z | 1.95 Å | 2021-12-22 | X-ray | FEY, FHS | MERS-CoV | NSP5 (3CLpro) | Functional ligand | 15.70% | 20.60% | 11.30% | (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[7-(phenylacetyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid | (1R,2S)-2-{[N-({[(2r,4R)-7-acetyl-7-azaspiro[3.5]non-5-en-2-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[7-(phenylacetyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid | XDS | 97.70% | 10.2 | APS (17-ID) | PHENIX | 2021-12-09 | 35638577 | - | - | - | - | N/A | - | - | 3C-like proteinase - Full Length | 81.7 | - | No | - | 77.3 | 93.3 | 78.5 | - | ||||||
| 7t40 | 1.70 Å | 2021-12-22 | X-ray | FVE, FV5, PG4 | MERS-CoV | NSP5 (3CLpro) | Functional ligand | 15.60% | 19.50% | 6.50% | (1R,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; (1S,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; (1R,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; TETRAETHYLENE GLYCOL | XDS | 99.50% | 10.5 | APS (17-ID) | PHENIX | 2021-12-09 | 35638577 | - | - | - | - | N/A | - | - | 3C-like proteinase - Full Length | 82.4 | - | No | - | 84.9 | 83.5 | 83.3 | - | ||||||
| 7t41 | 2.10 Å | 2021-12-22 | X-ray | FWI, FZI | MERS-CoV | NSP5 (3CLpro) | Functional ligand | 16.50% | 23.70% | 12.20% | (1R,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid | (1R,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid; (1S,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid | XDS | 97.60% | 7.9 | NSLS-II (19-ID) | PHENIX | 2021-12-09 | 35638577 | - | - | - | - | N/A | - | - | 3C-like proteinase - Full Length | 71.8 | - | No | - | 47.7 | 88.6 | 83.6 | - | ||||||
| 7t42 | 1.60 Å | 2021-12-22 | X-ray | FIK, PG4, FIW | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.00% | 21.20% | 8.80% | (1S,2S)-2-[(N-{[(2-acetyl-2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-[(N-{[(2-acetyl-2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; TETRAETHYLENE GLYCOL; (1R,2S)-1-hydroxy-2-{[N-({[2-(2-methylpropanoyl)-2-azaspiro[3.3]heptan-6-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | XDS | 99.70% | 9.7 | NSLS-II (19-ID) | PHENIX | 2021-12-09 | 35638577 | - | - | - | - | N/A | - | - | 3C-like proteinase | 71.3 | - | No | - | 72.3 | 57.3 | 88.9 | - | ||||||
| 7t43 | 1.70 Å | 2021-12-22 | X-ray | FN2, FP8, PG4 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.70% | 21.30% | 4.90% | (1S,2S)-1-hydroxy-2-[(N-{[(2-methyl-2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-1-hydroxy-2-[(N-{[(2-methyl-2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; (1R,2S)-1-hydroxy-2-[(N-{[(2-methyl-2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; TETRAETHYLENE GLYCOL | XDS | 97.60% | 9.9 | NSLS-II (19-ID) | PHENIX | 2021-12-09 | 35638577 | - | - | - | - | N/A | - | - | 3C-like proteinase | 79.9 | - | No | - | 71.4 | 80.8 | 90.3 | - | ||||||
| 7t44 | 1.45 Å | 2021-12-22 | X-ray | ET6, ESS, PG4 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.50% | 22.60% | 6.50% | (1R,2S)-2-[(N-{[(2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; (1S,2S)-1-hydroxy-2-{[N-({[2-(methanesulfonyl)-2-azaspiro[3.3]heptan-6-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-1-hydroxy-2-{[N-({[2-(methanesulfonyl)-2-azaspiro[3.3]heptan-6-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; (1R,2S)-2-[(N-{[(2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; TETRAETHYLENE GLYCOL | XDS | 98.70% | 10.0 | NSLS-II (19-ID) | PHENIX | 2021-12-09 | 35638577 | - | - | - | - | N/A | - | - | 3C-like proteinase | 61.4 | - | No | - | 59.1 | 49.6 | 88.7 | - | ||||||
| 7t45 | 1.65 Å | 2021-12-22 | X-ray | EW9, PG4 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.45% | 22.05% | 5.30% | (1S,2S)-2-{[N-({[7-(tert-butoxycarbonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-{[N-({[7-(tert-butoxycarbonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; TETRAETHYLENE GLYCOL | XDS | 99.60% | 11.1 | APS (17-ID) | PHENIX | 2021-12-09 | 35638577 | - | - | - | - | N/A | - | - | 3C-like proteinase | 61.2 | - | No | - | 64.6 | 51.3 | 81.0 | - | ||||||
| 7t46 | 1.45 Å | 2021-12-22 | X-ray | F8C, F5L | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.90% | 19.30% | 4.70% | (1S,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; (1R,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | XDS | 99.60% | 10.6 | APS (17-ID) | PHENIX | 2021-12-09 | 35638577 | - | - | - | - | N/A | - | - | 3C-like proteinase - Full Length | 77.1 | - | No | - | 86.1 | 68.2 | 79.4 | - | ||||||
| 7t48 | 1.90 Å | 2021-12-22 | X-ray | FHS, FEY | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.20% | 24.20% | 9.60% | (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[7-(phenylacetyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid | (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[7-(phenylacetyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid; (1R,2S)-2-{[N-({[(2r,4R)-7-acetyl-7-azaspiro[3.5]non-5-en-2-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | XDS | 99.00% | 8.4 | APS (17-ID) | PHENIX | 2021-12-09 | 35638577 | - | - | - | - | N/A | - | - | 3C-like proteinase - Full Length | 69.7 | - | No | - | 42.9 | 93.3 | 79.0 | - | ||||||
| 7t49 | 1.75 Å | 2021-12-22 | X-ray | FV5, FVE | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.00% | 24.10% | 5.90% | (1R,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1R,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; (1S,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | XDS | 98.90% | 10.1 | APS (17-ID) | PHENIX | 2021-12-09 | 35638577 | - | - | - | - | N/A | - | - | 3C-like proteinase - Full Length | 64.5 | - | No | - | 43.8 | 80.8 | 79.3 | - | ||||||
| 7t4a | 1.80 Å | 2021-12-22 | X-ray | PO4, EQS, EO6, PG4 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.40% | 22.50% | 5.00% | PHOSPHATE ION; (1S,2S)-2-[(N-{[(7-cyano-7-azaspiro[3.5]nonan-2-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; (1R,2S)-2-[(N-{[(7-cyano-7-azaspiro[3.5]nonan-2-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | PHOSPHATE ION; (1S,2S)-2-[(N-{[(7-cyano-7-azaspiro[3.5]nonan-2-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; (1R,2S)-2-[(N-{[(7-cyano-7-azaspiro[3.5]nonan-2-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; TETRAETHYLENE GLYCOL | XDS | 99.40% | 12.7 | APS (17-ID) | PHENIX | 2021-12-09 | 35638577 | - | - | - | - | N/A | - | - | 3C-like proteinase | 76.8 | - | No | - | 60.1 | 82.9 | 89.7 | - | ||||||
| 7t4b | 1.60 Å | 2021-12-22 | X-ray | FZI, FWI, PG4 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.50% | 21.60% | 5.80% | (1R,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid | (1S,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid; (1R,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid; TETRAETHYLENE GLYCOL | XDS | 99.20% | 11.5 | NSLS-II (19-ID) | PHENIX | 2021-12-09 | 35638577 | - | - | - | - | N/A | - | - | 3C-like proteinase - Full Length | 69.9 | - | No | - | 68.8 | 63.6 | 82.9 | - | ||||||
| 7vxb | 3.90 Å | 2021-12-22 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-11-12 | 34930910 | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7vxc | 3.90 Å | 2021-12-22 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-11-12 | 34930910 | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7vxi | 3.40 Å | 2021-12-22 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-11-12 | 34930910 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7vxk | 3.70 Å | 2021-12-22 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-11-12 | 34930910 | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7dqa | 2.80 Å | 2021-12-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2020-12-22 | - | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7dr8 | 2.34 Å | 2021-12-29 | X-ray | - | MERS-CoV | NSP5 (3CLpro) | No functional ligands | 21.36% | 26.70% | N/A | - | DENZO | 100.00% | 13.7 | SSRF (BL17U1) | PHENIX | 2020-12-26 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 16.4 | - | No | - | - | 21.7 | 27.8 | 56.6 | - | ||||||
| 7dr9 | 2.78 Å | 2021-12-29 | X-ray | - | MERS-CoV | NSP5 (3CLpro) | No functional ligands | 22.33% | 27.18% | N/A | - | AUTOMAR | 96.80% | 13.4 | SSRF (BL17U1) | PHENIX | 2020-12-27 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 19.0 | - | No | - | - | 18.6 | 46.0 | 47.7 | - | ||||||
| 7dra | 2.78 Å | 2021-12-29 | X-ray | - | MERS-CoV | NSP5 (3CLpro) | No functional ligands | 21.87% | 26.15% | N/A | - | CrysalisPro | 96.70% | 12.9 | SSRF (BL17U1) | PHENIX | 2020-12-27 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 27.5 | - | No | - | - | 25.5 | 57.4 | 47.6 | - | ||||||
| 7sxr | 3.00 Å | 2021-12-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-11-24 | 34914928 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7sxs | 2.64 Å | 2021-12-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-11-24 | 34914928 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7sxt | 2.31 Å | 2021-12-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-11-24 | 34914928 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7sxu | 3.00 Å | 2021-12-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-11-24 | 34914928 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7sxv | 2.79 Å | 2021-12-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-11-24 | 34914928 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7sxw | 2.85 Å | 2021-12-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-11-24 | 34914928 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7sxx | 2.66 Å | 2021-12-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-11-24 | 34914928 | - | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7sxy | 2.79 Å | 2021-12-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-11-24 | 34914928 | - | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7sxz | 2.61 Å | 2021-12-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-11-24 | 34914928 | - | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7sy0 | 3.00 Å | 2021-12-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-11-24 | 34914928 | - | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7sy1 | 2.83 Å | 2021-12-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-11-24 | 34914928 | - | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7sy2 | 3.11 Å | 2021-12-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-11-24 | 34914928 | - | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7sy3 | 2.95 Å | 2021-12-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-11-24 | 34914928 | - | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7sy4 | 3.35 Å | 2021-12-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-11-24 | 34914928 | - | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7sy5 | 2.59 Å | 2021-12-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-11-24 | 34914928 | - | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7sy6 | 2.81 Å | 2021-12-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-11-24 | 34914928 | - | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7sy7 | 2.81 Å | 2021-12-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-11-24 | 34914928 | - | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7sy8 | 3.14 Å | 2021-12-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-11-24 | 34914928 | - | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7t9j | 2.79 Å | 2021-12-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-12-19 | 35050643 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7t9k | 2.45 Å | 2021-12-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-12-19 | 35050643 | - | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7t9l | 2.66 Å | 2021-12-29 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-12-19 | 35050643 | - | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7t9w | 2.20 Å | 2021-12-29 | X-ray | - | SARS-CoV-2 | NSP3(C-term) | No functional ligands | 26.16% | 31.62% | 10.40% | - | HKL-3000 | 100.00% | 17.4 | ROTATING ANODE () | PHENIX | 2021-12-20 | - | - | - | - | - | N/A | - | - | Papain-like protease nsp3 - BSM (Betacoronavirus-Specific Marker) domain | 25.2 | - | No | - | - | 5.3 | 72.0 | 48.5 | - | ||||||
| 7spo | 1.92 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | Spike | Protein-protein complex | 19.04% | 22.63% | 4.60% | - | XDS | 99.90% | 19.6 | APS (24-ID-C) | PHENIX | 2021-11-02 | 34916516 | - | - | - | - | N/A | - | - | VNAR 3B4; Spike protein S1 | 50.2 | - | No | - | 58.8 | 35.9 | 80.8 | - | |||||||
| 7spp | 1.96 Å | 2022-01-05 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 18.80% | 22.43% | 5.60% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 98.70% | 11.4 | APS (24-ID-C) | PHENIX | 2021-11-02 | 34916516 | - | - | - | - | N/A | - | - | VNAR 2C02; Spike protein S1 | 42.2 | - | No | - | 60.9 | 15.0 | 84.3 | - | ||||||
| 7sjs | 1.61 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | Spike | Protein-protein complex | 17.39% | 20.63% | 7.90% | stem helix peptide of Spike protein S2' | - | HKL-2000 | 97.00% | 8.9 | SSRL (BL12-1) | PHENIX | 2021-11-02 | 35133175 | - | - | - | - | N/A | - | - | CC40.8 Fab light chain; CC40.8 Fab heavy chain; stem helix peptide of Spike protein S2' | 76.2 | - | No | - | 77.1 | 67.8 | 86.3 | - | ||||||
| 7tas | 3.20 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2021-11-02 | 34990214 | - | - | - | - | N/A | - | - | S2K146 Fab light chain; S2K146 Fab heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7tat | 3.20 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-11-02 | 34990214 | - | - | - | - | N/A | - | - | S2K146 Fab heavy chain; Spike glycoprotein; S2K146 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7tb4 | 3.29 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-02 | 33655252 | - | - | - | - | N/A | - | - | Surface glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7te0 | 2.00 Å | 2022-01-05 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.91% | 24.76% | N/A | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | PROTEUM PLUS | 94.60% | 15.9 | ROTATING ANODE () | PHENIX | 2021-11-02 | 35731933 | - | - | - | - | N/A | - | - | 3C-like proteinase | 28.9 | - | No | - | 37.6 | 28.0 | 68.0 | - | ||||||
| 7vxm | 3.60 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-11-02 | 34930910 | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7w92 | 3.10 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2021-11-02 | 35169135 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7w94 | 3.40 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2021-11-02 | 35169135 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7w98 | 3.60 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2021-11-02 | 35169135 | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7w99 | 3.40 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2021-11-02 | 35169135 | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7w9b | 3.40 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2021-11-02 | 35169135 | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7w9c | 3.20 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2021-11-02 | 35169135 | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7w9e | 3.10 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2021-11-02 | 35169135 | - | - | - | - | N/A | - | - | Anti-H5N1 hemagglutinin monoclonal anitbody H5M9 heavy chain; Spike glycoprotein; The light chain of 8D3 fab | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7w9f | 3.60 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2021-11-02 | 35169135 | - | - | - | - | N/A | - | - | The heavy chain of 8D3; Spike protein S1; The light chain of 8D3 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7w9i | 3.40 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2021-11-02 | 35169135 | - | - | - | - | N/A | - | - | Spike protein S1; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7dvp | 1.69 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.90% | 21.87% | 5.10% | nsp4/5 peptidyl substrate | - | XDS | 87.20% | 13.71 | SSRF (BL19U1) | PHENIX | 2021-11-02 | 35380892 | - | - | - | - | N/A | - | - | 3C-like proteinase; nsp4/5 peptidyl substrate | 61.3 | - | No | - | 66.4 | 44.0 | 86.8 | - | ||||||
| 7dvw | 1.49 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 13.95% | 17.26% | 4.90% | nsp5/6 peptidyl substrate | - | XDS | 97.80% | 19.72 | SSRF (BL19U1) | PHENIX | 2021-11-02 | 35380892 | - | - | - | - | N/A | - | - | nsp5/6 peptidyl substrate; 3C-like proteinase | 60.3 | - | No | - | 94.5 | 15.6 | 85.0 | - | ||||||
| 7dvx | 1.80 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.94% | 24.72% | 5.60% | nsp6/7 peptidyl substrate | - | XDS | 98.30% | 13.78 | SSRF (BL19U1) | PHENIX | 2021-11-02 | 35380892 | - | - | - | - | N/A | - | - | 3C-like proteinase; nsp6/7 peptidyl substrate | 36.4 | - | No | - | 37.9 | 38.7 | 72.4 | - | ||||||
| 7dvy | 1.80 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.51% | 21.63% | 6.80% | nsp9/10 peptidyl substrate | - | XDS | 95.30% | 10.91 | SSRF (BL19U1) | PHENIX | 2021-11-02 | 35380892 | - | - | - | - | N/A | - | - | 3C-like proteinase; nsp9/10 peptidyl substrate | 70.5 | - | No | - | 68.6 | 69.6 | 78.7 | - | ||||||
| 7dw0 | 1.81 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.45% | 20.77% | 5.80% | nsp15/16 peptidyl substrate | - | xia2 | 99.70% | 15.4 | SSRF (BL19U1) | PHENIX | 2021-11-02 | 35380892 | - | - | - | - | N/A | - | - | 3C-like proteinase; nsp15/16 peptidyl substrate | 49.0 | - | No | - | 76.0 | 31.6 | 65.5 | - | ||||||
| 7dw6 | 1.70 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.62% | 22.09% | 4.00% | nsp14/15 peptidyl substrate | - | XDS | 95.80% | 16.15 | SSRF (BL19U1) | PHENIX | 2021-11-02 | 35380892 | - | - | - | - | N/A | - | - | 3C-like proteinase; nsp14/15 peptidyl substrate | 43.0 | - | No | - | 64.4 | 19.4 | 78.0 | - | ||||||
| 7f5f | 1.62 Å | 2022-01-05 | X-ray | CA | SARS-CoV-2 | ORF8 | No functional ligands | 17.43% | 20.50% | N/A | CALCIUM ION | HKL-3000 | 100.00% | 58.9 | SSRF (BL19U1) | PHENIX | 2021-11-02 | 34975921 | CA | - | - | - | N/A | - | - | ORF8 protein | 63.3 | - | No | - | 78.0 | 23.5 | 99.7 | - | |||||||
| 7f8l | 1.76 Å | 2022-01-05 | X-ray | CA | Bat-CoV-RaTG13 | ORF8 | No functional ligands | 18.14% | 22.60% | N/A | CALCIUM ION | CALCIUM ION | 95.89% | 17.2 | SSRF (BL19U1) | PHENIX | 2021-11-02 | 34975921 | CA | - | - | - | N/A | - | - | Nonstructural protein NS8 | 40.6 | - | No | - | 59.1 | 46.4 | 51.5 | - | |||||||
| 7msq | 2.29 Å | 2022-01-05 | X-ray | SCN | SARS-CoV-2 | Spike | Protein-protein complex | 19.42% | 24.47% | 7.50% | THIOCYANATE ION | THIOCYANATE ION | XDS | 99.80% | 22.5 | Australian Synchrotron (MX2) | REFMAC | 2021-11-02 | 34788600 | - | - | - | - | N/A | - | - | Spike protein S1 - receptor binding domain; AB-3467 Fab Light Chain; AB-3467 Fab Heavy Chain | 28.0 | - | No | - | 40.5 | 35.0 | 56.2 | - | ||||||
| 7pki | 2.94 Å | 2022-01-05 | X-ray | NAG, TRS | cave bat sarbecovirus | Spike | Protein-protein complex | 21.44% | 22.72% | 5.00% | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL | XDS | 86.98% | 11.42 | SOLEIL (PROXIMA 1) | PHENIX | 2021-11-02 | 35172323 | ZN | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; BANAL 236 coronavirus spike receptor-binding domain | 70.8 | - | No | - | 57.9 | 85.2 | 74.2 | - | ||||||
| 7pku | - | 2022-01-05 | NMR | - | SARS-CoV-2 | NSP3(Ubl1)/Nucleocapsid | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2021-11-02 | 35044811 | - | - | - | - | N/A | - | - | Nucleoprotein; 3C-like proteinase | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7qnw | 2.40 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | Spike | Protein-protein complex | 20.56% | 24.74% | 33.90% | - | xia2 | 100.00% | 8.5 | Diamond (I03) | PHENIX | 2021-11-02 | 35081335 | - | - | - | - | N/A | - | - | EY6A light chain; Surface glycoprotein; Beta-55 heavy chain; Beta-55 light chain; EY6A heavy chain | 37.2 | - | No | - | 37.7 | 45.2 | 67.5 | - | |||||||
| 7qnx | 2.92 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | Spike | Protein-protein complex | 22.86% | 26.63% | 55.40% | - | xia2 | 99.90% | 3.8 | Diamond (I03) | PHENIX | 2021-11-02 | 35081335 | - | - | - | - | N/A | - | - | EY6A light chain; EY6A heavy chain; Beta-55 heavy chain; Beta-55 light chain; Surface glycoprotein | 28.2 | - | No | - | 22.0 | 32.4 | 77.8 | - | |||||||
| 7qny | 2.84 Å | 2022-01-05 | X-ray | NAG, NA | SARS-CoV-2 | Spike | Protein-protein complex | 22.64% | 27.83% | 29.00% | 2-acetamido-2-deoxy-beta-D-glucopyranose; SODIUM ION | 2-acetamido-2-deoxy-beta-D-glucopyranose; SODIUM ION | xia2 | 100.00% | 5.0 | Diamond (I03) | PHENIX | 2021-11-02 | 35081335 | - | - | - | - | N/A | - | - | COVOX-158 light chain; Surface glycoprotein; COVOX-158 heavy chain; COVOX-58 heavy chain; COVOX-58 light chain | 20.6 | - | No | - | 14.9 | 26.6 | 74.3 | - | ||||||
| 7qo7 | 3.02 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-02 | - | - | - | - | - | N/A | - | - | Surface glycoprotein,Fibritin,SARS-CoV-2 S Omicron Spike B.1.1.529 | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7thh | 1.32 Å | 2022-01-05 | X-ray | IOD, P6G | SARS-CoV-2 | NSP3(Cleavage_domain, PLpro) | No functional ligands | 14.46% | 19.09% | 13.60% | HEXAETHYLENE GLYCOL; IODIDE ION | IODIDE ION; HEXAETHYLENE GLYCOL | HKL-3000 | 99.30% | 6.3 | APS (19-ID) | REFMAC | 2021-11-02 | - | - | - | - | - | N/A | - | - | Papain-like protease nsp3 - SUD-C and Ubl2 domains, residues 1496-1623 | 80.5 | - | No | - | - | 87.3 | 71.4 | 85.8 | - | |||||
| 7vph | 2.80 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | ORF6 | Protein-protein complex | 19.24% | 24.42% | N/A | ORF6 protein | - | HKL-2000 | 99.32% | 1.0 | SSRF (BL17U1) | PHENIX | 2021-11-02 | 35096974 | - | - | - | - | N/A | - | - | mRNA export factor; ORF6 protein; Isoform 3 of Nuclear pore complex protein Nup98-Nup96 | 32.0 | - | No | - | 40.8 | 75.5 | 23.6 | - | ||||||
| 7wbl | 3.40 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-02 | 35093192 | ZN | - | - | - | N/A | - | - | Spike protein S1; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7wbp | 3.00 Å | 2022-01-05 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 18.09% | 20.37% | 19.00% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.90% | 10.6 | SSRF (BL02U1) | PHENIX | 2021-11-02 | 35093192 | ZN | - | - | - | N/A | - | - | Spike protein S1; Angiotensin-converting enzyme 2 | 77.0 | - | No | - | 79.1 | 75.5 | 78.7 | - | ||||||
| 7wbq | 3.34 Å | 2022-01-05 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 19.74% | 22.64% | 16.20% | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; beta-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 100.00% | 9.4 | SSRF (BL02U1) | PHENIX | 2021-11-02 | 35093192 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike protein S1 | 46.8 | - | No | - | 58.7 | 46.9 | 63.3 | - | ||||||
| 7dyd | 2.39 Å | 2022-01-05 | X-ray | EY3 | MERS-CoV | Nucleocapsid | Functional ligand | 25.78% | 29.52% | 9.10% | 5-propan-2-yloxy-1H-indole | 5-propan-2-yloxy-1H-indole | HKL-2000 | 98.80% | 7.8 | NSRRC (TPS 05A) | PHENIX | 2021-11-02 | 35517857 | - | - | - | - | N/A | - | - | Nucleoprotein | - | - | No | - | 8.4 | - | 25.5 | - | ||||||
| 7fcd | 3.90 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-11-02 | 35017654 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7fce | 3.10 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-11-02 | 35017654 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7nwx | 1.80 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 20.41% | 24.45% | 12.00% | - | XDS | 99.70% | 13.4 | SEALED TUBE () | PHENIX | 2021-11-02 | 34278402 | ZN | - | - | - | N/A | - | - | Replicase polyprotein 1a | 42.3 | - | No | - | 40.6 | 56.1 | 63.7 | - | |||||||
| 7nxh | 2.10 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 21.65% | 26.80% | 17.00% | - | XDS | 95.40% | 8.48 | SEALED TUBE () | PHENIX | 2021-11-02 | 34278402 | - | - | - | - | N/A | - | - | 3C-like proteinase | 16.7 | - | No | - | 20.8 | 20.3 | 65.8 | - | |||||||
| 7od3 | 2.80 Å | 2022-01-05 | Cryo-EM | EIC, NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | LINOLEIC ACID; 2-acetamido-2-deoxy-beta-D-glucopyranose | LINOLEIC ACID; 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-02 | 35017512 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7odl | 3.03 Å | 2022-01-05 | Cryo-EM | EIC, NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | LINOLEIC ACID; 2-acetamido-2-deoxy-beta-D-glucopyranose | LINOLEIC ACID; 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-02 | 35017512 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7qgi | 1.65 Å | 2022-01-05 | X-ray | PO4 | SARS-CoV-2 | NSP14 | No functional ligands | 19.71% | 21.99% | 12.50% | PHOSPHATE ION | PHOSPHATE ION | XDS | 89.20% | 9.2 | Diamond (I04-1) | PHENIX; PHENIX | 2021-11-02 | 36546776 | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 44.7 | - | No | - | 64.5 | 22.2 | 78.1 | - | ||||||
| 7qo9 | 3.88 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-02 | - | - | - | - | - | N/A | - | - | Spike glycoprotein,SARS-CoV-2 S Omicron Spike B.1.1.529 | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7ti9 | 2.73 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | NSP3: Ubl1 | No functional ligands | 22.21% | 24.86% | 6.30% | - | HKL-3000 | 98.00% | 29.2 | ROTATING ANODE () | PHENIX | 2021-11-02 | - | - | - | - | - | N/A | - | - | Papain-like protease nsp3 - ubiquitin-like domain 1 (Ubl1) | 37.0 | - | No | - | - | 36.6 | 71.4 | 42.0 | - | ||||||
| 7tll | 1.63 Å | 2022-01-05 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 21.11% | 24.98% | 5.60% | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | autoPROC | 78.00% | 11.5 | APS (17-ID) | BUSTER | 2021-11-02 | 35461811 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 42.7 | - | No | - | 35.5 | 38.3 | 87.2 | - | ||||||
| 7vjw | 2.20 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 22.41% | 28.15% | N/A | - | CrystFEL | 100.00% | 4.5 | FREE ELECTRON LASER () | PHENIX | 2021-11-02 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 20.7 | - | No | - | - | 13.4 | 57.3 | 45.4 | - | ||||||
| 7vjx | 2.20 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 25.57% | 28.68% | N/A | - | CrystFEL | 100.00% | 9.8 | FREE ELECTRON LASER () | PHENIX | 2021-11-02 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 20.4 | - | No | - | - | 11.2 | 31.6 | 72.7 | - | ||||||
| 7vjy | 1.90 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 23.29% | 27.84% | N/A | - | CrystFEL | 100.00% | 6.0 | FREE ELECTRON LASER () | PHENIX | 2021-11-02 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 3.2 | - | No | - | - | 14.9 | 11.7 | 34.4 | - | ||||||
| 7vjz | 1.90 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 21.73% | 25.00% | N/A | - | CrystFEL | 100.00% | 7.4 | FREE ELECTRON LASER () | PHENIX | 2021-11-02 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 26.4 | - | No | - | - | 35.1 | 11.3 | 81.9 | - | ||||||
| 7vk0 | 2.10 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 23.67% | 24.20% | N/A | - | CrystFEL | 100.00% | 7.5 | FREE ELECTRON LASER () | PHENIX | 2021-11-02 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 24.0 | - | No | - | - | 42.9 | 28.1 | 52.2 | - | ||||||
| 7vk1 | 1.93 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 21.69% | 26.49% | N/A | - | CrystFEL | 94.40% | 4.4 | FREE ELECTRON LASER () | PHENIX | 2021-11-02 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 15.0 | - | No | - | - | 23.2 | 18.3 | 61.2 | - | ||||||
| 7vk2 | 2.00 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 22.94% | 27.47% | N/A | - | CrystFEL | 100.00% | 7.6 | FREE ELECTRON LASER () | PHENIX | 2021-11-02 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 9.1 | - | No | - | - | 16.9 | 11.4 | 57.3 | - | ||||||
| 7vk3 | 2.10 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 23.78% | 27.80% | N/A | - | CrystFEL | 100.00% | 5.6 | FREE ELECTRON LASER () | PHENIX | 2021-11-02 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 13.5 | - | No | - | - | 15.0 | 29.7 | 54.1 | - | ||||||
| 7vk4 | 2.10 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 22.52% | 26.63% | N/A | - | CrystFEL | 100.00% | 10.8 | FREE ELECTRON LASER () | PHENIX | 2021-11-02 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 23.2 | - | No | - | - | 22.0 | 33.4 | 66.1 | - | ||||||
| 7vk5 | 2.17 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 24.07% | 26.97% | N/A | - | CrystFEL | 100.00% | 7.9 | FREE ELECTRON LASER () | PHENIX | 2021-11-02 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 15.7 | - | No | - | - | 19.8 | 28.1 | 56.5 | - | ||||||
| 7vk6 | 2.25 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 23.29% | 26.57% | N/A | - | CrystFEL | 100.00% | 8.6 | FREE ELECTRON LASER () | PHENIX | 2021-11-02 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 27.0 | - | No | - | - | 22.6 | 63.6 | 43.3 | - | ||||||
| 7vk7 | 2.40 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 20.89% | 23.95% | N/A | - | CrystFEL | 100.00% | 8.6 | FREE ELECTRON LASER () | PHENIX | 2021-11-02 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 45.4 | - | 49.9 | - | ||||||
| 7vk8 | 2.40 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 27.16% | 32.94% | N/A | - | CrystFEL | 99.40% | 1.9 | FREE ELECTRON LASER () | PHENIX | 2021-11-02 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 1.0 | - | No | - | - | 4.4 | 11.5 | 29.3 | - | ||||||
| 7wev | 3.60 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2021-11-02 | 34930910 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7wk2 | 3.10 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-11-02 | 35228716 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wk3 | 3.40 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-11-02 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7wk8 | 3.61 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-11-02 | 35228716 | - | - | - | - | N/A | - | - | Heavy chain of S3H3 Fab; Light chain of S3H3 Fab; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wka | 3.64 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-11-02 | 35228716 | - | - | - | - | N/A | - | - | Spike glycoprotein; Heavy chain of S3H3 Fab; Light chain of S3H3 Fab | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7f7e | 2.49 Å | 2022-01-05 | X-ray | NAG | SARS-CoV-2 | Spike | Protein-protein complex | 21.44% | 25.62% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 96.03% | 14.22 | APS (24-ID-E) | PHENIX | 2021-11-02 | 35060840 | - | - | - | - | N/A | - | - | Spike protein S1 - RBD domain; Heavy chain of A5-10 Fab; Light chain of A5-10 Fab | 4.5 | - | No | - | 29.7 | 12.4 | 25.5 | - | ||||||
| 7nll | 2.89 Å | 2022-01-05 | Cryo-EM | RTV | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-(acetylamino)-1,5-anhydro-2-deoxy-D-mannitol | 2-(acetylamino)-1,5-anhydro-2-deoxy-D-mannitol | - | - | () | PHENIX | 2021-11-02 | 35013189 | - | - | - | - | N/A | - | - | Surface glycoprotein; Nanobody Fu2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7ns6 | 3.18 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-02 | 35013189 | - | - | - | - | N/A | - | - | Spike glycoprotein,Fibritin; Fu2 nanobody | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7nt4 | 2.68 Å | 2022-01-05 | X-ray | PRL | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 19.32% | 26.41% | N/A | PROFLAVIN | PROFLAVIN | XDS | 95.70% | 13.2 | SLS (X06DA) | REFMAC | 2021-11-02 | 35021060 | ZN | - | - | - | N/A | - | - | Non-structural protein 3 | 10.8 | - | No | - | 23.6 | 52.9 | 14.6 | - | ||||||
| 7pqy | 3.00 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | Spike | Protein-protein complex | 22.79% | 24.46% | 18.10% | - | xia2 | 99.40% | 7.2 | Diamond (I03) | PHENIX | 2021-11-02 | 34987630 | - | - | - | - | N/A | - | - | FI-3A Fab light chain; Spike protein S1; FI-3A Fab heavy chain | 25.0 | - | No | - | 40.5 | 18.7 | 66.2 | - | |||||||
| 7pqz | 3.20 Å | 2022-01-05 | X-ray | PO4 | SARS-CoV-2 | Spike | Protein-protein complex | 20.06% | 24.01% | 26.30% | PHOSPHATE ION | PHOSPHATE ION | xia2 | 100.00% | 13.6 | Diamond (I03) | PHENIX | 2021-11-02 | 34987630 | - | - | - | - | N/A | - | - | FD-11A Fab heavy chain; FD-11A Fab light chain; Spike protein S1; FI-3A Fab heavy chain; FI-3A Fab light chain | 24.9 | - | No | - | 44.7 | 13.2 | 67.3 | - | ||||||
| 7pr0 | 2.92 Å | 2022-01-05 | X-ray | NO3, NAG | SARS-CoV-2 | Spike | Protein-protein complex | 21.36% | 25.06% | 28.40% | NITRATE ION; 2-acetamido-2-deoxy-beta-D-glucopyranose | NITRATE ION; 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 99.80% | 5.7 | Diamond (I03) | PHENIX | 2021-11-02 | 34987630 | - | - | - | - | N/A | - | - | Spike protein S1; FD-5D Fab heavy chain; FD-5D Fab light chain | 25.1 | - | No | - | 34.7 | 20.3 | 70.7 | - | ||||||
| 7qif | 2.53 Å | 2022-01-05 | X-ray | GTG, PO4 | SARS-CoV-2 | NSP14 | Functional ligand | 20.44% | 24.17% | 58.50% | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE; PHOSPHATE ION | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE; PHOSPHATE ION | DIALS | 91.70% | 6.0 | Diamond (I03) | PHENIX | 2021-11-02 | 36546776 | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | - | - | No | - | 38.7 | - | 83.8 | - | ||||||
| 7sc1 | 3.20 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-02 | 34973165 | - | - | - | - | N/A | - | - | R40-1G8 Fab light chain; Spike glycoprotein; R40-1G8 Fab heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tla | 3.13 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2021-11-02 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7tlb | 3.06 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2021-11-02 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7tlc | 2.83 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2021-11-02 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7tld | 2.89 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2021-11-02 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7tly | 3.00 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2021-11-02 | 35076256 | - | - | - | - | N/A | - | - | S309 Fab heavy chain; Spike glycoprotein; S309 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7tlz | 3.30 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-11-02 | 35076256 | - | - | - | - | N/A | - | - | Spike glycoprotein; S2L20 Fab light chain; S2L20 Fab heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7tm0 | 3.10 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-02 | 35076256 | - | - | - | - | N/A | - | - | S309 Fab light chain; S2L20 Fab heavy chain; S2L20 Fab light chain; Spike glycoprotein; S309 Fab Heavy Chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tn0 | 2.85 Å | 2022-01-05 | X-ray | NAG | SARS-CoV-2 | Spike | No functional ligands | 23.03% | 26.79% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 96.70% | 5.6 | ALS (4.2.2) | PHENIX; PHENIX | 2021-11-02 | 35076256 | ZN | - | - | - | N/A | - | - | Spike protein S1 - Receptor-binding domain; S304 Fab heavy chain; S304 Fab light chain; Angiotensin-converting enzyme 2; S309 heavy chain; S309 light chain | 47.7 | - | No | - | 21.1 | 74.0 | 75.8 | - | |||||||
| 7tob | 2.05 Å | 2022-01-05 | X-ray | K36 | SARS-CoV-2 | NSP5 (3CLpro) | Protein-protein complex | 17.16% | 21.35% | 7.70% | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | 99.20% | 9.4 | ROTATING ANODE () | REFMAC | 2021-11-02 | 35292745 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 56.2 | - | No | - | 71.1 | 48.9 | 67.0 | - | |||||||
| 7wcr | 3.50 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2021-11-02 | 34964428 | - | - | - | - | N/A | - | - | Light chain of S5D2 Fab; Heavy chain of S5D2 Fab; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7wcz | 3.50 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2021-11-02 | 34964428 | - | - | - | - | N/A | - | - | Heavy chain of S5D2 Fab; Light chain of S5D2 Fab; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7wd0 | 3.30 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2021-11-02 | 34964428 | - | - | - | - | N/A | - | - | Spike glycoprotein; Heavy chain of S5D2 Fab; Light chain of S5D2 Fab | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7wd7 | 3.50 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2021-11-02 | 34964428 | - | - | - | - | N/A | - | - | Light chain of S5D2 Fab; Spike glycoprotein; Heavy chain of S5D2 Fab | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7wd8 | 4.30 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2021-11-02 | 34964428 | - | - | - | - | N/A | - | - | Heavy chain of S3H3 Fab; Light chain of S3H3 Fab; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7wd9 | 3.70 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2021-11-02 | 34964428 | - | - | - | - | N/A | - | - | Heavy chain of S3H3 Fab; Spike glycoprotein; Light chain of S3H3 Fab | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7wdf | 3.90 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2021-11-02 | 34964428 | - | - | - | - | N/A | - | - | Heavy chain of S3H3 Fab; Light chain of S3H3 Fab; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7wk4 | 3.69 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-11-02 | - | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7wk5 | 3.66 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | 2021-11-02 | - | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7wk6 | 3.67 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-11-02 | - | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7wk9 | 3.48 Å | 2022-01-05 | Cryo-EM | - | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-11-02 | 35228716 | - | - | - | - | N/A | - | - | Heavy chain of S3H3 Fab; Spike glycoprotein; Light chain of S3H3 Fab | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 5sbf | 1.64 Å | 2022-01-05 | X-ray | - | SARS-CoV-2 | NSP15 | PanDDA fragment screening | 18.75% | 21.11% | 9.00% | - | XDS | 99.90% | 12.2 | Diamond (I04) | REFMAC | 2021-11-02 | 37115000 | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease | - | - | No | - | 73.1 | - | - | - | |||||||
| 7lhq | - | 2022-01-05 | NMR | - | SARS-CoV-2 | NSP7 | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2021-11-02 | - | - | - | - | - | N/A | - | - | Non-structural protein 7 | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7tou | 3.24 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-02 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tov | 3.16 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-02 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tox | 3.62 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-02 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7toy | 3.53 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-02 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7toz | 4.07 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-02 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tp0 | 3.57 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-02 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tp1 | 3.81 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-02 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tp2 | 3.72 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-02 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tp7 | 3.48 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-02 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tp8 | 3.40 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-02 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tp9 | 3.48 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-02 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tpa | 3.60 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-02 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tpc | 3.91 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-02 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tpe | 4.00 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-02 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tpf | 3.40 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-02 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tph | 3.58 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-02 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tpl | 3.87 Å | 2022-01-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-02 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tq5 | 1.65 Å | 2022-01-05 | X-ray | IRW, ITX, PG4 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.90% | 21.70% | 5.40% | (1R,2S)-1-hydroxy-2-{[N-({[(1R,2R)-2-(4-methoxyphenyl)cyclopropyl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; (1S,2S)-1-hydroxy-2-{[N-({[(1R,2R)-2-(4-methoxyphenyl)cyclopropyl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-1-hydroxy-2-{[N-({[(1R,2R)-2-(4-methoxyphenyl)cyclopropyl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; (1R,2S)-1-hydroxy-2-{[N-({[(1R,2R)-2-(4-methoxyphenyl)cyclopropyl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; TETRAETHYLENE GLYCOL | XDS | 99.90% | 11.2 | APS (17-ID) | PHENIX | 2021-11-02 | 36654747 | - | - | - | - | N/A | - | - | 3C-like proteinase | 71.3 | - | No | - | 67.9 | 65.9 | 84.8 | - | ||||||
| 7tq6 | 1.55 Å | 2022-01-05 | X-ray | IT3, ITG, PG4 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.50% | 21.60% | 4.20% | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid; (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid | (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid; (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid; TETRAETHYLENE GLYCOL | XDS | 99.90% | 13.2 | APS (17-ID) | PHENIX | 2021-11-02 | 36654747 | - | - | - | - | N/A | - | - | 3C-like proteinase - Full Length | 64.2 | - | No | - | 68.8 | 46.4 | 87.9 | - | ||||||
| 7tq7 | 1.70 Å | 2022-01-05 | X-ray | PG4, IRR | MERS-CoV | NSP5 (3CLpro) | Functional ligand | 15.93% | 19.02% | 6.00% | N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucinamide | TETRAETHYLENE GLYCOL; N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucinamide | XDS | 99.60% | 11.3 | APS (17-ID) | PHENIX | 2021-11-02 | 36654747 | - | - | - | - | N/A | - | - | Orf1a protein | 85.5 | - | No | - | 87.6 | 88.6 | 92.2 | - | ||||||
| 7ed5 | 2.98 Å | 2022-02-16 | Cryo-EM | AT9 | SARS-CoV-2 | NSP7/NSP8/NSP12 | Functional ligand | N/A | N/A | N/A | RNA (5'-R(P*GP*CP*UP*AP*UP*GP*UP*GP*AP*GP*AP*UP*UP*AP*AP*GP*UP*UP*AP*U*(AT9))-3'); RNA (5'-R(P*CP*CP*CP*CP*AP*UP*AP*AP*CP*UP*UP*AP*AP*UP*CP*UP*CP*AP*CP*AP*UP*AP*GP*C)-3'); [[(2R,3R,4R,5R)-5-(2-azanyl-6-oxidanylidene-1H-purin-9-yl)-4-fluoranyl-4-methyl-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | [[(2R,3R,4R,5R)-5-(2-azanyl-6-oxidanylidene-1H-purin-9-yl)-4-fluoranyl-4-methyl-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | - | - | () | PHENIX | 2021-03-15 | 35110538 | MG; ZN | - | - | - | N/A | - | - | Non-structural protein 8; RNA-directed RNA polymerase; Non-structural protein 7; RNA (5'-R(P*CP*CP*CP*CP*AP*UP*AP*AP*CP*UP*UP*AP*AP*UP*CP*UP*CP*AP*CP*AP*UP*AP*GP*C)-3'); RNA (5'-R(P*GP*CP*UP*AP*UP*GP*UP*GP*AP*GP*AP*UP*UP*AP*AP*GP*UP*UP*AP*U*(AT9))-3') | N/A | - | No | - | N/A | N/A | N/A | RNA (5'-R(P*CP*CP*CP*CP*AP*UP*AP*AP*CP*UP*UP*AP*AP*UP*CP*UP*CP*AP*CP*AP*UP*AP*GP*C)-3'); RNA (5'-R(P*GP*CP*UP*AP*UP*GP*UP*GP*AP*GP*AP*UP*UP*AP*AP*GP*UP*UP*AP*U*(AT9))-3') | - | ||||||
| 7tei | 3.40 Å | 2022-02-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-05 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tf8 | 3.36 Å | 2022-02-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-06 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7the | 3.79 Å | 2022-02-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-01-10 | 35447081 | - | - | - | - | N/A | - | - | DH1042 Fab Light Chain; DH1042 Fab Heavy Chain; Spike protein S1 - receptor binding domain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tht | 3.42 Å | 2022-02-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-12 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein; DH1042 light chain; DH1042 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tnw | 3.10 Å | 2022-02-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-21 | 35452593 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7to4 | 3.40 Å | 2022-02-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-22 | 35452593 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tow | 2.15 Å | 2022-02-16 | X-ray | PO4, CA | SARS-CoV-2 | Spike | Protein-protein complex | 16.68% | 23.37% | N/A | Spike protein S2; PHOSPHATE ION; CALCIUM ION | PHOSPHATE ION; CALCIUM ION | HKL-2000 | 92.90% | 22.5 | APS (22-ID) | PHENIX | 2022-01-24 | 35447081 | CA | - | - | - | N/A | - | - | Spike protein S2; DH1058 Fab Light chain; DH1058 Fab heavy chain | 46.2 | - | No | - | 51.4 | 71.2 | 45.1 | - | ||||||
| 7tvs | 1.89 Å | 2022-02-16 | X-ray | XNJ | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.93% | 22.49% | 9.80% | N-(4-methyl-3-{[4-(pyridin-3-yl)-1,3-thiazol-2-yl]amino}phenyl)-4-[(piperazin-1-yl)methyl]benzamide | N-(4-methyl-3-{[4-(pyridin-3-yl)-1,3-thiazol-2-yl]amino}phenyl)-4-[(piperazin-1-yl)methyl]benzamide | HKL-3000 | 98.30% | 17.6 | APS (19-ID) | PHENIX | 2022-02-05 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 58.9 | - | No | - | - | 60.4 | 55.5 | 76.4 | - | |||||
| 7tvx | 2.09 Å | 2022-02-16 | X-ray | G65 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.83% | 21.18% | 8.50% | Masitinib | HKL-3000 | 95.80% | 15.6 | APS (19-ID) | PHENIX | 2022-02-06 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 73.7 | - | No | - | - | 72.7 | 55.4 | 96.4 | - | ||||||
| 7au4 | 1.82 Å | 2022-02-23 | X-ray | RY5 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.49% | 21.39% | 5.60% | (3~{S})-6-chloranyl-3'-(1,2-oxazol-3-ylmethyl)spiro[1,2-dihydroindene-3,5'-imidazolidine]-2',4'-dione | (3~{S})-6-chloranyl-3'-(1,2-oxazol-3-ylmethyl)spiro[1,2-dihydroindene-3,5'-imidazolidine]-2',4'-dione | XDS | 99.80% | 8.4 | MAX IV (BioMAX) | PHENIX | 2020-11-02 | 35142215 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 73.5 | - | No | - | 70.8 | 78.7 | 74.5 | - | ||||||
| 7b2j | 1.55 Å | 2022-02-23 | X-ray | SQ2 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.15% | 20.26% | 6.70% | 2-(1H-1,2,3-benzotriazol-1-yl)-1-(4-methylpiperidin-1-yl)ethan-1-one | 2-(1H-1,2,3-benzotriazol-1-yl)-1-(4-methylpiperidin-1-yl)ethan-1-one | XDS | 97.80% | 13.2 | MAX IV (BioMAX) | PHENIX; REFMAC | 2020-11-27 | 35142215 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 81.3 | - | No | - | 79.9 | 78.7 | 88.7 | - | ||||||
| 7b2u | 1.55 Å | 2022-02-23 | X-ray | SQ5 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.83% | 24.63% | 6.00% | (5S)-5-(cyclohexylmethyl)-3-(5-fluoropyridin-3-yl)imidazolidine-2,4-dione | (5S)-5-(cyclohexylmethyl)-3-(5-fluoropyridin-3-yl)imidazolidine-2,4-dione | XDS | 96.50% | 10.1 | MAX IV (BioMAX) | PHENIX; REFMAC | 2020-11-27 | 35142215 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 50.8 | - | No | - | 38.8 | 54.1 | 83.7 | - | ||||||
| 7b5z | 1.65 Å | 2022-02-23 | X-ray | SYH | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 15.54% | 19.61% | 5.80% | 2-(1H-benzo[d][1,2,3]triazol-1-yl)-1-(4-methylenepiperidin-1-yl)ethan-1-one | 2-(1H-benzo[d][1,2,3]triazol-1-yl)-1-(4-methylenepiperidin-1-yl)ethan-1-one | XDS | 98.60% | 18.6 | MAX IV (BioMAX) | PHENIX; REFMAC | 2020-12-07 | 35142215 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 78.2 | - | No | - | 84.2 | 68.7 | 83.9 | - | ||||||
| 7b77 | 1.60 Å | 2022-02-23 | X-ray | T0W | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.86% | 21.38% | 5.50% | 2-(benzotriazol-1-yl)-~{N}-ethyl-~{N}-(furan-3-ylmethyl)ethanamide | 2-(benzotriazol-1-yl)-~{N}-ethyl-~{N}-(furan-3-ylmethyl)ethanamide | XDS | 99.30% | 15.0 | MAX IV (BioMAX) | PHENIX; REFMAC | 2020-12-09 | 35142215 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 75.5 | - | No | - | 70.9 | 64.3 | 93.9 | - | ||||||
| 7bij | 1.47 Å | 2022-02-23 | X-ray | TU8 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.54% | 21.25% | 5.80% | (3~{S})-3'-(5-fluoranylpyridin-3-yl)spiro[1,2-dihydroindene-3,5'-imidazolidine]-2',4'-dione | (3~{S})-3'-(5-fluoranylpyridin-3-yl)spiro[1,2-dihydroindene-3,5'-imidazolidine]-2',4'-dione | XDS | 98.50% | 13.7 | MAX IV (BioMAX) | REFMAC; REFMAC | 2021-01-12 | 35142215 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 66.8 | - | No | - | 72.0 | 52.2 | 84.4 | - | ||||||
| 7e6k | 1.60 Å | 2022-02-23 | X-ray | HYR | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.04% | 20.18% | 5.20% | N-(2-phenoxyethyl)methanethioamide | N-(2-phenoxyethyl)methanethioamide | 99.70% | 18.1 | SSRF (BL17U1) | PHENIX | 2021-02-22 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 49.8 | - | No | - | - | 80.5 | 29.4 | 64.7 | - | ||||||
| 7nbt | 1.63 Å | 2022-02-23 | X-ray | U7W | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.04% | 23.02% | 6.00% | 2-(benzotriazol-1-yl)-1-[(4~{S})-4-methyl-6,7-dihydro-4~{H}-thieno[3,2-c]pyridin-5-yl]ethanone | 2-(benzotriazol-1-yl)-1-[(4~{S})-4-methyl-6,7-dihydro-4~{H}-thieno[3,2-c]pyridin-5-yl]ethanone | XDS | 98.10% | 11.1 | MAX IV (BioMAX) | REFMAC | 2021-01-27 | 35142215 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 65.0 | - | No | - | 54.8 | 64.3 | 85.8 | - | ||||||
| 7neo | 1.64 Å | 2022-02-23 | X-ray | U9H | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.12% | 23.87% | 6.50% | 2-cyclobutyl-7-(5-fluoropyridin-3-yl)-5,7-diazaspiro[3.4]octane-6,8-dione | 2-cyclobutyl-7-(5-fluoropyridin-3-yl)-5,7-diazaspiro[3.4]octane-6,8-dione | XDS | 99.90% | 11.3 | MAX IV (BioMAX) | REFMAC | 2021-02-04 | 35142215 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 56.9 | - | No | - | 46.2 | 60.1 | 82.0 | - | ||||||
| 7o46 | 2.23 Å | 2022-02-23 | X-ray | V18 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.58% | 24.87% | 5.50% | 2-cyclobutyl-7-isoquinolin-4-yl-5,7-diazaspiro[3.4]octane-6,8-dione | 2-cyclobutyl-7-isoquinolin-4-yl-5,7-diazaspiro[3.4]octane-6,8-dione | XDS | 99.70% | 14.6 | MAX IV (BioMAX) | REFMAC | 2021-04-05 | 35142215 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 47.1 | - | No | - | 36.5 | 51.8 | 81.2 | - | ||||||
| 7q0a | 4.80 Å | 2022-02-23 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-10-14 | 34987630 | - | - | - | - | N/A | - | - | Spike glycoprotein,Spike ectodomain,Spike protein S2'; FI3A fab heavy chain; FI3A fab Light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7qbb | 2.00 Å | 2022-02-23 | X-ray | V1B | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.70% | 25.80% | 7.00% | 7-isoquinolin-4-yl-2-phenyl-5,7-diazaspiro[3.4]octane-6,8-dione | 7-isoquinolin-4-yl-2-phenyl-5,7-diazaspiro[3.4]octane-6,8-dione | XDS | 99.70% | 11.9 | MAX IV (BioMAX) | REFMAC | 2021-11-18 | 35142215 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 36.0 | - | No | - | 28.1 | 48.6 | 71.4 | - | ||||||
| 7tl1 | 3.50 Å | 2022-02-23 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-18 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7tl9 | 3.50 Å | 2022-02-23 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-18 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tp3 | 2.33 Å | 2022-02-23 | X-ray | CAC | SARS-CoV-2 | Spike | Protein-protein complex | 18.52% | 23.06% | 14.30% | CACODYLATE ION | CACODYLATE ION | HKL-2000 | 97.70% | 7.6 | SSRL (BL12-1) | PHENIX | 2022-01-24 | 35947674 | - | - | - | - | N/A | - | - | K288.2 heavy chain; K288.2 light chain; Spike protein S1 | 35.8 | - | No | - | 54.5 | 37.9 | 55.4 | - | ||||||
| 7tp4 | 1.95 Å | 2022-02-23 | X-ray | - | SARS-CoV-2 | Spike | Protein-protein complex | 20.46% | 23.06% | 9.00% | - | autoPROC | 99.80% | 12.1 | APS (23-ID-D) | PHENIX | 2022-01-24 | 35947674 | - | - | - | - | N/A | - | - | Spike protein S1 - Receptor binding domain, UNP residues 333-530; K398.22 light chain; K398.22 heavy chain | 31.1 | - | No | - | 54.5 | 28.6 | 55.1 | - | |||||||
| 7twf | 1.10 Å | 2022-02-23 | X-ray | - | SARS-CoV-2 | NSP3: Macro | No functional ligands | 11.31% | 13.46% | 6.39% | - | XDS | 99.86% | 11.83 | SSRL (BL12-1) | PHENIX | 2022-02-07 | 35622909 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain (UNP residues 1025-1191) | 86.3 | - | No | - | 99.4 | 80.5 | 95.7 | - | |||||||
| 7twg | 1.10 Å | 2022-02-23 | X-ray | - | SARS-CoV-2 | NSP3: Macro | No functional ligands | 10.54% | 12.63% | 5.29% | - | Adxv | 99.70% | 14.17 | SSRL (BL12-1) | PHENIX | 2022-02-07 | 35622909 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain (UNP residues 1025-1191) | 86.8 | - | No | - | 99.7 | 84.9 | 97.8 | - | |||||||
| 7twh | 1.10 Å | 2022-02-23 | X-ray | - | SARS-CoV-2 | NSP3: Macro | No functional ligands | 9.94% | 11.47% | 3.40% | - | XDS | 99.77% | 21.87 | SSRL (BL12-1) | PHENIX | 2022-02-07 | 35622909 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain (UNP residues 1025-1191) | 86.5 | - | No | - | 99.9 | 80.5 | 97.1 | - | |||||||
| 7twi | 1.10 Å | 2022-02-23 | X-ray | - | SARS-CoV-2 | NSP3: Macro | No functional ligands | 9.56% | 11.34% | 3.10% | - | XDS | 99.45% | 25.61 | SSRL (BL12-1) | PHENIX | 2022-02-07 | 35622909 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain (UNP residues 1025-1191) | 85.1 | - | No | - | 99.9 | 71.4 | 94.7 | - | |||||||
| 7twj | 0.90 Å | 2022-02-23 | X-ray | - | SARS-CoV-2 | NSP3: Macro | No functional ligands | 9.69% | 10.92% | 2.48% | - | XDS | 99.90% | 33.33 | ALS (8.3.1) | PHENIX | 2022-02-07 | 35622909 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 84.8 | - | No | - | 99.9 | 80.5 | 83.8 | - | |||||||
| 7twn | 0.90 Å | 2022-02-23 | X-ray | NHE | SARS-CoV-2 | NSP3: Macro | No functional ligands | 9.93% | 11.35% | 2.68% | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID | XDS | 99.95% | 32.28 | ALS (8.3.1) | PHENIX | 2022-02-07 | 35622909 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 86.0 | - | No | - | 99.9 | 89.6 | 83.3 | - | ||||||
| 7two | 0.90 Å | 2022-02-23 | X-ray | NHE | SARS-CoV-2 | NSP3: Macro | No functional ligands | 10.46% | 11.87% | 2.73% | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID | XDS | 99.78% | 28.5 | ALS (8.3.1) | PHENIX | 2022-02-07 | 35622909 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 81.9 | - | No | - | 99.8 | 67.2 | 82.7 | - | ||||||
| 7twp | 0.90 Å | 2022-02-23 | X-ray | - | SARS-CoV-2 | NSP3: Macro | No functional ligands | 10.83% | 12.40% | 3.02% | - | XDS | 99.87% | 25.98 | ALS (8.3.1) | PHENIX | 2022-02-07 | 35622909 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 85.6 | - | No | - | 99.7 | 75.9 | 93.9 | - | |||||||
| 7twq | 0.90 Å | 2022-02-23 | X-ray | - | SARS-CoV-2 | NSP3: Macro | No functional ligands | 10.96% | 12.58% | 2.70% | - | XDS | 99.89% | 28.95 | ALS (8.3.1) | PHENIX | 2022-02-07 | 35622909 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 85.2 | - | No | - | 99.7 | 71.6 | 95.4 | - | |||||||
| 7twr | 0.90 Å | 2022-02-23 | X-ray | - | SARS-CoV-2 | NSP3: Macro | No functional ligands | 10.60% | 12.12% | 2.99% | - | XDS | 99.73% | 26.06 | ALS (8.3.1) | PHENIX | 2022-02-07 | 35622909 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 85.5 | - | No | - | 99.8 | 76.0 | 93.1 | - | |||||||
| 7tws | 0.90 Å | 2022-02-23 | X-ray | - | SARS-CoV-2 | NSP3: Macro | No functional ligands | 11.12% | 12.65% | 2.52% | - | XDS | 99.91% | 30.16 | ALS (8.3.1) | PHENIX | 2022-02-07 | 35622909 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 84.3 | - | No | - | 99.7 | 67.2 | 94.7 | - | |||||||
| 7twt | 0.90 Å | 2022-02-23 | X-ray | AR6 | SARS-CoV-2 | NSP3: Macro | Functional ligand | 10.63% | 12.02% | 4.75% | [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | XDS | 99.84% | 17.44 | ALS (8.3.1) | PHENIX | 2022-02-07 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 | 86.4 | - | No | - | - | 99.8 | 80.5 | 96.3 | - | |||||
| 7twv | 0.90 Å | 2022-02-23 | X-ray | AR6 | SARS-CoV-2 | NSP3: Macro | Functional ligand | 9.50% | 10.66% | 2.33% | [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | XDS | 99.88% | 40.56 | ALS (8.3.1) | PHENIX | 2022-02-07 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 | 85.9 | - | No | - | - | 99.9 | 85.1 | 86.7 | - | |||||
| 7tww | 0.90 Å | 2022-02-23 | X-ray | AR6 | SARS-CoV-2 | NSP3: Macro | Functional ligand | 9.44% | 10.68% | 2.50% | [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | XDS | 99.85% | 36.47 | ALS (8.3.1) | PHENIX | 2022-02-07 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 | 84.4 | - | No | - | - | 99.9 | 76.2 | 85.6 | - | |||||
| 7twx | 0.90 Å | 2022-02-23 | X-ray | AR6 | SARS-CoV-2 | NSP3: Macro | Functional ligand | 9.54% | 10.86% | 2.63% | [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | XDS | 99.89% | 36.03 | ALS (8.3.1) | PHENIX | 2022-02-07 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 | 83.6 | - | No | - | - | 99.9 | 76.0 | 81.3 | - | |||||
| 7twy | 0.90 Å | 2022-02-23 | X-ray | AR6 | SARS-CoV-2 | NSP3: Macro | Functional ligand | 9.30% | 10.67% | 2.18% | [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | XDS | 99.89% | 40.54 | ALS (8.3.1) | PHENIX | 2022-02-07 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 | 86.3 | - | No | - | - | 99.9 | 89.7 | 85.4 | - | |||||
| 7tx0 | 0.84 Å | 2022-02-23 | X-ray | AR6 | SARS-CoV-2 | NSP3: Macro | Functional ligand | 10.00% | 11.10% | 2.84% | [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | XDS | 99.90% | 41.39 | ALS (8.3.1) | PHENIX | 2022-02-07 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 | 86.1 | - | No | - | - | 99.9 | 80.4 | 92.8 | - | |||||
| 7tx1 | 0.90 Å | 2022-02-23 | X-ray | AR6 | SARS-CoV-2 | NSP3: Macro | Functional ligand | 9.59% | 10.77% | 2.43% | [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | XDS | 99.90% | 35.99 | ALS (8.3.1) | PHENIX | 2022-02-07 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 | 85.3 | - | No | - | - | 99.9 | 80.5 | 86.8 | - | |||||
| 7tx3 | 1.60 Å | 2022-02-23 | NEUTRONDIFFRACTION/X-RAY/HYBRID | NHE | SARS-CoV-2 | NSP3: Macro | Functional ligand | 12.39% | 15.76% | 8.70% | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID | Mantid | 99.60% | 15.6 | ROTATING ANODE, SPALLATION SOURCE () | PHENIX | 2022-02-07 | 35622909 | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||
| 7tx4 | 1.90 Å | 2022-02-23 | NEUTRONDIFFRACTION/X-RAY/HYBRID | - | SARS-CoV-2 | NSP3: Macro | No functional ligands | 16.62% | 22.40% | 9.70% | - | Mantid | 96.60% | 16.9 | SPALLATION SOURCE, ROTATING ANODE () | PHENIX | 2022-02-07 | 35622909 | - | - | - | - | N/A | - | - | Papain-like protease nsp3 - macrodomain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tx5 | 1.95 Å | 2022-02-23 | NEUTRONDIFFRACTION/X-RAY/HYBRID | APR | SARS-CoV-2 | NSP3: Macro | Functional ligand | 13.22% | 17.19% | 7.00% | ADENOSINE-5-DIPHOSPHORIBOSE | ADENOSINE-5-DIPHOSPHORIBOSE | 92.90% | 19.2 | SPALLATION SOURCE, ROTATING ANODE () | PHENIX | 2022-02-07 | 35622909 | - | - | - | - | N/A | - | - | Papain-like protease nsp3 - macrodomain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7e5x | 2.19 Å | 2022-03-02 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 23.47% | 26.61% | 3.70% | - | HKL-2000 | 97.10% | 16.94 | SSRF (BL19U1) | REFMAC | 2021-02-21 | 35380892 | - | - | - | - | N/A | - | - | 3C-like proteinase | 9.7 | - | No | - | 22.1 | 14.9 | 50.5 | - | |||||||
| 7nij | 1.58 Å | 2022-03-02 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 17.71% | 20.31% | 7.00% | - | XDS | 99.40% | 9.2 | ESRF (ID23-2) | PHENIX | 2021-02-12 | 35234150 | - | - | - | - | N/A | - | - | 2'-O-methyltransferase | 66.7 | - | No | - | 79.5 | 39.3 | 89.7 | - | |||||||
| 7ntq | 1.50 Å | 2022-03-02 | X-ray | NA, 35J, FMT | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.06% | 21.32% | N/A | FORMIC ACID; SODIUM ION; N-(pyridin-3-ylmethyl)thioformamide | SODIUM ION; N-(pyridin-3-ylmethyl)thioformamide; FORMIC ACID | Aimless | 98.70% | 9.77 | SOLEIL (PROXIMA 1) | REFMAC | 2021-03-10 | 36796300 | - | - | - | - | N/A | - | - | 3C-like proteinase | 75.1 | - | No | - | 71.3 | 73.6 | 83.2 | - | ||||||
| 7ntt | 1.74 Å | 2022-03-02 | X-ray | NA, FMT | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 20.73% | 25.47% | N/A | SODIUM ION; FORMIC ACID | SODIUM ION; FORMIC ACID | Aimless | 95.00% | 7.72 | SOLEIL (PROXIMA 1) | REFMAC | 2021-03-10 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 42.9 | - | No | - | - | 31.1 | 58.1 | 72.4 | - | |||||
| 7ntw | 1.81 Å | 2022-03-02 | X-ray | NA, FMT | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 19.66% | 25.70% | N/A | SODIUM ION; FORMIC ACID | SODIUM ION; FORMIC ACID | Aimless | 99.00% | 12.8 | SOLEIL (PROXIMA 1) | REFMAC | 2021-03-11 | - | NA; ZN | - | - | - | N/A | - | - | 3C-like proteinase | 40.8 | - | No | - | - | 29.0 | 64.3 | 64.2 | - | |||||
| 7r10 | 4.00 Å | 2022-03-02 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-02 | 35246509 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7r15 | 4.10 Å | 2022-03-02 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-02 | 35246509 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7r1a | 3.90 Å | 2022-03-02 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-02 | 35246509 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7skq | 3.16 Å | 2022-03-02 | X-ray | TTT | BtSCoV-Rf1.2004 | NSP3: PLpro | Functional ligand | 23.02% | 25.91% | N/A | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide | 100.00% | 11.6 | NSLS-II (17-ID-1) | PHENIX | 2021-10-21 | 35199517 | ZN | - | - | - | N/A | - | - | 3C-like proteinase | 48.4 | - | No | - | 27.2 | 86.3 | 58.5 | - | |||||||
| 7skr | 2.89 Å | 2022-03-02 | X-ray | 9OZ | BtSCoV-Rf1.2004 | NSP3: PLpro | Functional ligand | 20.09% | 23.99% | N/A | N-[(2-methoxypyridin-4-yl)methyl]-2-[(1R)-1-(naphthalen-1-yl)ethyl]-2-azaspiro[3.3]heptane-6-carboxamide | N-[(2-methoxypyridin-4-yl)methyl]-2-[(1R)-1-(naphthalen-1-yl)ethyl]-2-azaspiro[3.3]heptane-6-carboxamide | HKL-2000 | 99.50% | 15.5 | APS (22-ID) | PHENIX | 2021-10-21 | 35199517 | ZN | - | - | - | N/A | - | - | 3C-like proteinase | 71.1 | - | No | - | 45.1 | 88.9 | 84.3 | - | ||||||
| 7tdu | 1.85 Å | 2022-03-02 | NEUTRONDIFFRACTION/X-RAY/HYBRID | I1W, DOD | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.70% | 21.00% | 7.60% | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxo(1-~2~H)pyrrolidin-3-yl]propan-2-yl}-3-{N-[tert-butyl(~2~H)carbamoyl]-3-methyl-L-(N-~2~H)valyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-(~2~H)carboxamide; water | CrysalisPro; LAUEGEN | 96.60% | 13.6 | ROTATING ANODE, NUCLEAR REACTOR () | nCNS | 2022-01-03 | 35477935 | - | - | - | - | N/A | - | - | 3C-like proteinase | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7teh | 1.80 Å | 2022-03-02 | X-ray | I1Z | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 15.51% | 18.56% | 3.70% | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | CrysalisPro | 93.60% | 33.35 | ROTATING ANODE () | PHENIX | 2022-01-05 | 35477935 | - | - | - | - | N/A | - | - | 3C-like proteinase | 59.0 | - | No | - | 89.9 | 42.8 | 59.8 | - | ||||||
| 7tfr | 1.80 Å | 2022-03-02 | X-ray | NB2 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 15.79% | 18.11% | 4.60% | (1R,2S,5S)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-({1-[(2-methylpropane-2-sulfonyl)methyl]cyclohexyl}carbamoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-({1-[(2-methylpropane-2-sulfonyl)methyl]cyclohexyl}carbamoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | CrysalisPro | 99.90% | 25.92 | ROTATING ANODE () | PHENIX | 2022-01-07 | 35477935 | - | - | - | - | N/A | - | - | 3C-like proteinase | 57.8 | - | No | - | 91.7 | 20.3 | 78.2 | - | ||||||
| 7thk | 3.11 Å | 2022-03-02 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-01-11 | 35172173 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7tq8 | 1.65 Å | 2022-03-02 | X-ray | PG4, IUB, IRK | MERS-CoV | NSP5 (3CLpro) | Functional ligand | 15.90% | 19.10% | 8.50% | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2S)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid; (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2S)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid | TETRAETHYLENE GLYCOL; (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2S)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid; (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2S)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid | XDS | 98.20% | 10.1 | APS (17-ID) | PHENIX | 2022-01-26 | 36654747 | - | - | - | - | N/A | - | - | Orf1a protein | 84.6 | - | No | - | 87.2 | 83.5 | 92.4 | - | ||||||
| 7tzj | 2.66 Å | 2022-03-02 | X-ray | S88 | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 20.06% | 25.74% | 6.17% | N-[(3-fluorophenyl)methyl]-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide | N-[(3-fluorophenyl)methyl]-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide | XDS | 99.80% | 7.01 | Australian Synchrotron (MX2) | PHENIX | 2022-02-15 | 35494659 | ZN | - | - | - | N/A | - | - | Papain-like protease | 38.6 | - | No | - | 28.8 | 91.0 | 33.4 | - | ||||||
| 7vvt | 2.51 Å | 2022-03-02 | X-ray | 80X | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 23.39% | 25.27% | 32.70% | N-(3-chlorophenyl)-2-[(2R)-1-ethanoyl-3-oxidanylidene-piperazin-2-yl]ethanamide | N-(3-chlorophenyl)-2-[(2R)-1-ethanoyl-3-oxidanylidene-piperazin-2-yl]ethanamide | HKL-3000 | 99.20% | 1.6 | SSRF (BL18U1) | PHENIX | 2021-11-08 | 35114541 | - | - | - | - | N/A | - | - | 3C-like proteinase | 45.7 | - | No | - | 32.9 | 36.0 | 98.1 | - | ||||||
| 7vyr | 2.20 Å | 2022-03-02 | X-ray | - | SARS-CoV-2 | Spike | NLF | 19.20% | 23.53% | 8.70% | - | HKL-2000 | 95.40% | 1.36 | PAL/PLS (11C) | PHENIX | 2021-11-15 | 35030983 | - | - | - | - | N/A | - | - | Spike protein S1; D27 light chain; D27 heavy chain | 37.6 | - | No | - | 49.6 | 33.6 | 68.0 | - | |||||||
| 7wvn | 3.40 Å | 2022-03-02 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-02-10 | 35228716 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wvo | 3.41 Å | 2022-03-02 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-02-10 | 35228716 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ywr | - | 2022-03-02 | NMR | - | SARS-CoV-2 | NSP8 | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2022-02-14 | - | - | - | - | - | N/A | - | - | ORF1a polyprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7e9o | 3.41 Å | 2022-03-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-03-04 | - | - | - | - | - | N/A | - | - | Light chain of 35B5 Fab; Heavy chain of 35B5 Fab; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7e9q | 3.65 Å | 2022-03-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-03-04 | - | - | - | - | - | N/A | - | - | Light chain of 35B5 Fab; Spike glycoprotein; Heavy chain of 35B5 Fab | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7q1z | 3.40 Å | 2022-03-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-10-22 | 35233549 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7r0z | 3.50 Å | 2022-03-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-02 | 35246509 | ZN | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7r11 | 3.50 Å | 2022-03-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-02 | 35246509 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7r12 | 3.30 Å | 2022-03-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-02 | 35246509 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7r13 | 3.60 Å | 2022-03-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-02 | 35246509 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7r14 | 3.40 Å | 2022-03-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-02 | 35246509 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7r16 | 3.50 Å | 2022-03-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-02 | 35246509 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7r17 | 3.70 Å | 2022-03-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-02 | 35246509 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7r18 | 3.00 Å | 2022-03-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-02 | 35246509 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7r19 | 3.30 Å | 2022-03-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-02 | 35246509 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7r1b | 2.80 Å | 2022-03-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-02 | 35246509 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7r2v | 2.53 Å | 2022-03-09 | X-ray | SAH, TRS | SARS-CoV-2 | NSP14 | No functional ligands | 19.63% | 25.43% | 16.00% | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL; S-ADENOSYL-L-HOMOCYSTEINE | S-ADENOSYL-L-HOMOCYSTEINE; 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL | XDS | 87.80% | 6.8 | NSLS (X6A) | REFMAC | 2022-02-06 | 35609600 | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 27.8 | - | No | - | 31.4 | 50.8 | 49.0 | - | ||||||
| 7s5p | 2.86 Å | 2022-03-09 | X-ray | NAG | SARS-CoV-2 | Spike | No functional ligands | 20.83% | 24.71% | 25.60% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 100.00% | 2.3 | ALS (5.0.1) | PHENIX | 2021-09-11 | 35076281 | - | - | - | - | N/A | - | - | Spike protein S1; CS23 Light chain; CS23 Heavy chain | 22.4 | - | No | - | 38.0 | 41.6 | 40.2 | - | ||||||
| 7s5q | 2.88 Å | 2022-03-09 | X-ray | NAG | SARS-CoV-2 | Spike | No functional ligands | 21.61% | 25.73% | 16.90% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 98.90% | 4.6 | SSRL (BL12-1) | PHENIX | 2021-09-11 | 35076281 | - | - | - | - | N/A | - | - | CS44 Light chain; COVA1-16 Heavy chain; COVA1-16 Light chain; CS44 Heavy chain; Spike protein S1 | 16.7 | - | No | - | 28.8 | 34.0 | 44.1 | - | ||||||
| 7s5r | 2.45 Å | 2022-03-09 | X-ray | NAG | SARS-CoV-2 | Spike | No functional ligands | 20.71% | 23.75% | 11.50% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.90% | 5.2 | SSRL (BL12-1) | PHENIX | 2021-09-11 | 35076281 | - | - | - | - | N/A | - | - | CV07-287 Heavy chain; CV07-287 Light chain; COVA1-16 Light chain; COVA1-16 Heavy chain; Spike protein S1 | 30.8 | - | No | - | 47.4 | 40.7 | 49.6 | - | ||||||
| 7tge | 3.68 Å | 2022-03-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-07 | 35447081 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7tgw | 3.00 Å | 2022-03-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-01-09 | 35241675 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7tgx | 3.20 Å | 2022-03-09 | Cryo-EM | MAN, NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | alpha-D-mannopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | alpha-D-mannopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-01-09 | 35241675 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7tgy | 3.00 Å | 2022-03-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-01-09 | 35241675 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7u28 | 1.68 Å | 2022-03-09 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.80% | 24.76% | 6.40% | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | autoPROC | 74.20% | 10.5 | APS (17-ID) | BUSTER | 2022-02-23 | 35461811 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 54.4 | - | No | - | 37.6 | 55.7 | 90.2 | - | ||||||
| 7u29 | 2.09 Å | 2022-03-09 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.41% | 26.55% | 11.70% | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | autoPROC | 85.70% | 7.9 | APS (17-ID) | BUSTER | 2022-02-23 | 35461811 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 52.2 | - | No | - | 22.6 | 78.7 | 77.9 | - | ||||||
| 7wp9 | 2.56 Å | 2022-03-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-01-23 | 35133176 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wpa | 2.77 Å | 2022-03-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-01-23 | 35133176 | ZN | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wpb | 2.79 Å | 2022-03-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-01-23 | 35133176 | ZN | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wpc | 2.57 Å | 2022-03-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-01-23 | 35133176 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7x08 | 2.70 Å | 2022-03-09 | Cryo-EM | NAG, EIC | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; LINOLEIC ACID | 2-acetamido-2-deoxy-beta-D-glucopyranose; LINOLEIC ACID | - | - | () | 2022-02-21 | 35169121 | - | - | - | - | N/A | - | - | Spike glycoprotein; heavy chain of 2G1; light chain of 2G1 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 5skw | 2.09 Å | 2022-03-16 | X-ray | PO4, LF6 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 19.70% | 26.71% | N/A | {(1R,2R)-2-[(Z)-(3-methyl-1,2,4-thiadiazol-5(2H)-ylidene)amino]cyclopentyl}methanol; PHOSPHATE ION | PHOSPHATE ION; {(1R,2R)-2-[(Z)-(3-methyl-1,2,4-thiadiazol-5(2H)-ylidene)amino]cyclopentyl}methanol | XDS | 91.70% | 7.0 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 25.9 | - | No | - | - | 21.4 | 43.7 | 62.1 | - | |||||
| 5skx | 2.34 Å | 2022-03-16 | X-ray | PO4, T6J | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 18.29% | 27.51% | N/A | PHOSPHATE ION; 2-[(methylsulfonyl)methyl]-1H-benzimidazole | PHOSPHATE ION; 2-[(methylsulfonyl)methyl]-1H-benzimidazole | XDS | 92.60% | 7.2 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 17.2 | - | No | - | - | 16.6 | 47.8 | 43.7 | - | |||||
| 5sky | 2.25 Å | 2022-03-16 | X-ray | PO4, O2M | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 21.33% | 24.83% | 23.70% | PHOSPHATE ION; N-[(4-methyl-1,3-thiazol-2-yl)methyl]-1H-pyrazole-5-carboxamide | PHOSPHATE ION; N-[(4-methyl-1,3-thiazol-2-yl)methyl]-1H-pyrazole-5-carboxamide | XDS | 100.00% | 6.4 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 33.7 | - | No | - | - | 36.8 | 40.0 | 66.7 | - | |||||
| 5skz | 1.96 Å | 2022-03-16 | X-ray | PO4, NVD | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 21.49% | 24.40% | 12.70% | PHOSPHATE ION; N-{[4-(dimethylamino)phenyl]methyl}-4H-1,2,4-triazol-4-amine | PHOSPHATE ION; N-{[4-(dimethylamino)phenyl]methyl}-4H-1,2,4-triazol-4-amine | XDS | 98.60% | 9.3 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 32.2 | - | No | - | - | 41.0 | 28.0 | 71.4 | - | |||||
| 5sl0 | 2.00 Å | 2022-03-16 | X-ray | PO4, B0V | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 22.38% | 25.97% | 11.90% | PHOSPHATE ION; 2-methoxy-~{N}-(2,4,6-trimethylphenyl)ethanamide | PHOSPHATE ION; 2-methoxy-~{N}-(2,4,6-trimethylphenyl)ethanamide | XDS | 97.70% | 10.4 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 27.2 | - | No | - | - | 26.9 | 31.2 | 72.0 | - | |||||
| 5sl1 | 2.38 Å | 2022-03-16 | X-ray | PO4, O2A | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 20.49% | 26.66% | 23.00% | PHOSPHATE ION; N-methyl-1H-indole-7-carboxamide | PHOSPHATE ION; N-methyl-1H-indole-7-carboxamide | XDS | 99.20% | 8.0 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 15.1 | - | No | - | - | 21.9 | 23.1 | 57.9 | - | |||||
| 5sl2 | 1.74 Å | 2022-03-16 | X-ray | PO4, LFO | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 22.59% | 25.63% | 17.80% | N,1-dimethyl-1H-indole-3-carboxamide; PHOSPHATE ION | PHOSPHATE ION; N,1-dimethyl-1H-indole-3-carboxamide | XDS | 99.80% | 7.6 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 30.1 | - | No | - | - | 29.6 | 24.6 | 81.9 | - | |||||
| 5sl3 | 1.99 Å | 2022-03-16 | X-ray | LGR, PO4 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 24.04% | 26.88% | 31.00% | PHOSPHATE ION; 2-[acetyl(methyl)amino]benzoic acid | 2-[acetyl(methyl)amino]benzoic acid; PHOSPHATE ION | XDS | 99.90% | 5.9 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 20.6 | - | No | - | - | 20.4 | 25.3 | 70.2 | - | |||||
| 5sl4 | 1.94 Å | 2022-03-16 | X-ray | PO4, LHR | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 24.29% | 28.09% | 21.90% | N-(1H-indazol-6-yl)acetamide; PHOSPHATE ION | PHOSPHATE ION; N-(1H-indazol-6-yl)acetamide | XDS | 99.50% | 6.6 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 15.8 | - | No | - | - | 13.7 | 25.3 | 65.7 | - | |||||
| 5sl5 | 2.36 Å | 2022-03-16 | X-ray | PO4, WNV | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 22.23% | 28.06% | 34.90% | N,N,2,3-tetramethylbenzamide; PHOSPHATE ION | PHOSPHATE ION; N,N,2,3-tetramethylbenzamide | XDS | 99.70% | 4.9 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 26.9 | - | No | - | - | 13.8 | 45.7 | 69.8 | - | |||||
| 5sl6 | 2.29 Å | 2022-03-16 | X-ray | PO4, 60P | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 23.49% | 29.29% | 31.80% | 3-methylthiophene-2-carboxylic acid; PHOSPHATE ION | PHOSPHATE ION; 3-methylthiophene-2-carboxylic acid | XDS | 99.90% | 2.8 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 10.1 | - | No | - | - | 9.1 | 37.9 | 41.5 | - | |||||
| 5sl7 | 1.84 Å | 2022-03-16 | X-ray | PO4, W0G | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 22.62% | 25.07% | 18.70% | (3R)-1-(2-fluorophenyl)-3-(methylamino)pyrrolidin-2-one; PHOSPHATE ION | PHOSPHATE ION; (3R)-1-(2-fluorophenyl)-3-(methylamino)pyrrolidin-2-one | XDS | 100.00% | 7.7 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 31.7 | - | No | - | - | 34.6 | 24.5 | 80.3 | - | |||||
| 5sl8 | 2.07 Å | 2022-03-16 | X-ray | PO4, JGD | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 22.70% | 26.11% | 34.60% | N,N-dimethylpyridin-4-amine; PHOSPHATE ION | PHOSPHATE ION; N,N-dimethylpyridin-4-amine | XDS | 99.70% | 6.4 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 20.5 | - | No | - | - | 25.7 | 15.2 | 74.6 | - | |||||
| 5sl9 | 1.75 Å | 2022-03-16 | X-ray | O0S, PO4 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 27.13% | 29.47% | 18.30% | N-{4-[(pyrimidin-2-yl)oxy]phenyl}acetamide; PHOSPHATE ION | N-{4-[(pyrimidin-2-yl)oxy]phenyl}acetamide; PHOSPHATE ION | XDS | 99.90% | 6.5 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 16.1 | - | No | - | - | 8.6 | 20.1 | 76.8 | - | |||||
| 5sla | 1.70 Å | 2022-03-16 | X-ray | LJR, PO4 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 25.11% | 27.14% | 13.30% | 1-cyclohexyl-N-methylmethanesulfonamide; PHOSPHATE ION | 1-cyclohexyl-N-methylmethanesulfonamide; PHOSPHATE ION | XDS | 99.90% | 8.1 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 16.9 | - | No | - | - | 18.7 | 21.7 | 66.8 | - | |||||
| 5slb | 1.80 Å | 2022-03-16 | X-ray | PO4, LJK | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 22.52% | 24.19% | 14.30% | 3-methyl-N-(2-methylbutan-2-yl)-1H-pyrazole-5-carboxamide; PHOSPHATE ION | PHOSPHATE ION; 3-methyl-N-(2-methylbutan-2-yl)-1H-pyrazole-5-carboxamide | XDS | 99.90% | 9.3 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 37.5 | - | No | - | - | 43.2 | 24.5 | 83.3 | - | |||||
| 5slc | 1.67 Å | 2022-03-16 | X-ray | U1V, PO4 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 24.19% | 25.96% | 12.30% | PHOSPHATE ION; 1-(4-fluoro-2-methylphenyl)methanesulfonamide | 1-(4-fluoro-2-methylphenyl)methanesulfonamide; PHOSPHATE ION | XDS | 99.50% | 9.5 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 26.6 | - | No | - | - | 26.9 | 21.7 | 80.0 | - | |||||
| 5sld | 1.58 Å | 2022-03-16 | X-ray | LJ6, PO4 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 24.53% | 25.83% | 10.40% | PHOSPHATE ION; (2R)-3-(3,5-dimethyl-1,2-oxazol-4-yl)-N,N,2-trimethylpropanamide | (2R)-3-(3,5-dimethyl-1,2-oxazol-4-yl)-N,N,2-trimethylpropanamide; PHOSPHATE ION | XDS | 99.40% | 10.7 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 24.7 | - | No | - | - | 28.0 | 20.1 | 76.8 | - | |||||
| 5sle | 2.01 Å | 2022-03-16 | X-ray | JGA, PO4 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 22.64% | 26.12% | 20.70% | N-ethyl-N'-(5-methyl-1,2-oxazol-3-yl)urea; PHOSPHATE ION | N-ethyl-N'-(5-methyl-1,2-oxazol-3-yl)urea; PHOSPHATE ION | XDS | 99.20% | 7.1 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 24.0 | - | No | - | - | 25.7 | 33.8 | 63.8 | - | |||||
| 5slf | 2.01 Å | 2022-03-16 | X-ray | LJA, PO4 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 20.17% | 24.51% | 23.60% | PHOSPHATE ION; N-[3-(carbamoylamino)phenyl]acetamide | N-[3-(carbamoylamino)phenyl]acetamide; PHOSPHATE ION | XDS | 99.90% | 6.6 | Diamond (I04-1) | REFMAC | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 22.5 | - | No | - | - | 39.9 | 36.9 | 43.2 | - | |||||
| 5slg | 1.97 Å | 2022-03-16 | X-ray | NZJ, PO4 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 27.44% | 32.34% | 29.30% | PHOSPHATE ION; 1-(3-methylbenzene-1-carbonyl)piperidine-4-carboxamide | 1-(3-methylbenzene-1-carbonyl)piperidine-4-carboxamide; PHOSPHATE ION | XDS | 98.80% | 4.2 | Diamond (I04-1) | REFMAC | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 4.9 | - | No | - | - | 4.8 | 26.2 | 38.6 | - | |||||
| 5slh | 1.82 Å | 2022-03-16 | X-ray | LLU, PO4 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 23.83% | 26.40% | 19.00% | PHOSPHATE ION; (2S)-2-(2-fluorophenoxy)propanoic acid | (2S)-2-(2-fluorophenoxy)propanoic acid; PHOSPHATE ION | XDS | 99.90% | 7.3 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 23.4 | - | No | - | - | 23.6 | 24.5 | 74.0 | - | |||||
| 5sli | 2.30 Å | 2022-03-16 | X-ray | PO4, LL0 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 20.81% | 27.13% | 27.00% | PHOSPHATE ION; 2-(difluoromethoxy)benzene-1-sulfonamide | PHOSPHATE ION; 2-(difluoromethoxy)benzene-1-sulfonamide | XDS | 100.00% | 4.7 | Diamond (I04-1) | REFMAC | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 11.5 | - | No | - | - | 18.8 | 37.9 | 36.5 | - | |||||
| 5slj | 2.31 Å | 2022-03-16 | X-ray | LKU, PO4 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 18.06% | 24.67% | 14.80% | PHOSPHATE ION; 3-fluoro-N-(3-hydroxy-4-methylphenyl)benzamide | 3-fluoro-N-(3-hydroxy-4-methylphenyl)benzamide; PHOSPHATE ION | XDS | 100.00% | 7.7 | Diamond (I04-1) | REFMAC | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 29.8 | - | No | - | - | 38.5 | 52.6 | 44.3 | - | |||||
| 5slk | 2.21 Å | 2022-03-16 | X-ray | PO4, LKL | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 22.03% | 28.46% | 27.40% | PHOSPHATE ION; 2-[(5-chloro-3-fluoropyridin-2-yl)(methyl)amino]ethan-1-ol | PHOSPHATE ION; 2-[(5-chloro-3-fluoropyridin-2-yl)(methyl)amino]ethan-1-ol | XDS | 99.90% | 5.2 | Diamond (I04-1) | REFMAC | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 11.3 | - | No | - | - | 12.1 | 38.3 | 42.1 | - | |||||
| 5sll | 1.81 Å | 2022-03-16 | X-ray | PO4, LK6 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 22.55% | 25.09% | 17.40% | PHOSPHATE ION; N-[(3R)-3-methyl-1,1-dioxo-1lambda~6~-thiolan-3-yl]cyclopropanecarboxamide | PHOSPHATE ION; N-[(3R)-3-methyl-1,1-dioxo-1lambda~6~-thiolan-3-yl]cyclopropanecarboxamide | XDS | 99.70% | 7.4 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 23.8 | - | No | - | - | 34.5 | 24.5 | 63.8 | - | |||||
| 5slm | 2.05 Å | 2022-03-16 | X-ray | WN1, PO4 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 25.82% | 30.72% | 31.10% | PHOSPHATE ION; N-(2-fluorophenyl)-3-methoxybenzamide | N-(2-fluorophenyl)-3-methoxybenzamide; PHOSPHATE ION | XDS | 100.00% | 5.0 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 12.4 | - | No | - | - | 6.2 | 22.4 | 66.8 | - | |||||
| 5sln | 2.21 Å | 2022-03-16 | X-ray | LUY, PO4 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 24.00% | 29.83% | 29.00% | PHOSPHATE ION; ~{N}-(2-phenylethyl)-1~{H}-benzimidazol-2-amine | ~{N}-(2-phenylethyl)-1~{H}-benzimidazol-2-amine; PHOSPHATE ION | XDS | 99.90% | 5.1 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 9.6 | - | No | - | - | 7.6 | 14.9 | 64.6 | - | |||||
| 5slo | 1.83 Å | 2022-03-16 | X-ray | JJM, PO4 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 24.32% | 28.10% | 20.40% | PHOSPHATE ION; 1-methyl-N-(3-methylphenyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine | 1-methyl-N-(3-methylphenyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine; PHOSPHATE ION | XDS | 99.80% | 7.1 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 21.0 | - | No | - | - | 13.5 | 26.5 | 76.6 | - | |||||
| 5slp | 1.82 Å | 2022-03-16 | X-ray | PO4, UWY | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 22.98% | 26.00% | 17.90% | PHOSPHATE ION; N-(1-ethyl-1H-pyrazol-4-yl)cyclopentanecarboxamide | PHOSPHATE ION; N-(1-ethyl-1H-pyrazol-4-yl)cyclopentanecarboxamide | XDS | 99.90% | 7.3 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 21.9 | - | No | - | - | 26.5 | 15.5 | 76.6 | - | |||||
| 5slq | 2.11 Å | 2022-03-16 | X-ray | PO4, ELQ | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 23.64% | 27.38% | 40.60% | PHOSPHATE ION; [3,4-bis(fluoranyl)phenyl]-(4-methylpiperazin-1-yl)methanone | PHOSPHATE ION; [3,4-bis(fluoranyl)phenyl]-(4-methylpiperazin-1-yl)methanone | XDS | 99.80% | 5.4 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 13.8 | - | No | - | - | 17.4 | 16.3 | 65.8 | - | |||||
| 5slr | 1.86 Å | 2022-03-16 | X-ray | LO6, PO4 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 23.64% | 26.72% | 19.50% | PHOSPHATE ION; 2-(difluoromethoxy)-1-[(2R,6S)-2,6-dimethylmorpholin-4-yl]ethan-1-one | 2-(difluoromethoxy)-1-[(2R,6S)-2,6-dimethylmorpholin-4-yl]ethan-1-one; PHOSPHATE ION | XDS | 99.90% | 7.0 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 16.2 | - | No | - | - | 21.4 | 12.6 | 71.6 | - | |||||
| 5sls | 2.29 Å | 2022-03-16 | X-ray | PO4, SZE | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 23.64% | 29.25% | 21.50% | 4-(3-fluoranylpyridin-2-yl)-1-methyl-piperazin-2-one; PHOSPHATE ION | PHOSPHATE ION; 4-(3-fluoranylpyridin-2-yl)-1-methyl-piperazin-2-one | XDS | 100.00% | 6.6 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 6.5 | - | No | - | - | 9.2 | 17.2 | 49.8 | - | |||||
| 5slt | 1.90 Å | 2022-03-16 | X-ray | PO4, LNS | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 22.65% | 25.70% | 16.50% | PHOSPHATE ION; 6-(methylcarbamoyl)pyridine-2-carboxylic acid | PHOSPHATE ION; 6-(methylcarbamoyl)pyridine-2-carboxylic acid | XDS | 99.90% | 8.2 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 27.2 | - | No | - | - | 29.0 | 25.3 | 75.8 | - | |||||
| 5slu | 2.09 Å | 2022-03-16 | X-ray | PO4, UX1 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 24.34% | 28.34% | 31.70% | 1-[(2-fluorophenyl)methyl]-N-methylcyclopropane-1-carboxamide; PHOSPHATE ION | PHOSPHATE ION; 1-[(2-fluorophenyl)methyl]-N-methylcyclopropane-1-carboxamide | XDS | 99.80% | 4.2 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 20.3 | - | No | - | - | 12.6 | 41.6 | 60.9 | - | |||||
| 5slv | 2.05 Å | 2022-03-16 | X-ray | EJQ, PO4 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 24.44% | 28.03% | 27.90% | PHOSPHATE ION; ~{N}-(4-fluorophenyl)-2-pyrrolidin-1-yl-ethanamide | ~{N}-(4-fluorophenyl)-2-pyrrolidin-1-yl-ethanamide; PHOSPHATE ION | XDS | 99.90% | 5.4 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 11.3 | - | No | - | - | 13.9 | 12.8 | 65.7 | - | |||||
| 5slw | 2.05 Å | 2022-03-16 | X-ray | PO4, U0V | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 23.76% | 27.34% | 29.00% | 2-fluoro-N-[2-(pyridin-4-yl)ethyl]benzamide; PHOSPHATE ION | PHOSPHATE ION; 2-fluoro-N-[2-(pyridin-4-yl)ethyl]benzamide | XDS | 99.60% | 5.8 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 22.3 | - | No | - | - | 17.6 | 29.0 | 73.0 | - | |||||
| 5slx | 1.76 Å | 2022-03-16 | X-ray | PO4, LMW | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 23.58% | 25.88% | 16.50% | 2-[(4-aminophenyl)(ethyl)amino]ethan-1-ol; PHOSPHATE ION | PHOSPHATE ION; 2-[(4-aminophenyl)(ethyl)amino]ethan-1-ol | XDS | 99.80% | 7.8 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 26.1 | - | No | - | - | 27.7 | 24.2 | 75.9 | - | |||||
| 5sly | 2.02 Å | 2022-03-16 | X-ray | PO4, LM6 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 25.47% | 29.23% | 29.00% | PHOSPHATE ION; 1-(1-ethyl-1H-pyrazol-5-yl)-N-methylmethanamine | PHOSPHATE ION; 1-(1-ethyl-1H-pyrazol-5-yl)-N-methylmethanamine | XDS | 99.80% | 5.3 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 18.3 | - | No | - | - | 9.2 | 32.4 | 68.9 | - | |||||
| 5slz | 2.54 Å | 2022-03-16 | X-ray | PO4, LQP | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 22.23% | 29.36% | 39.60% | 2-(difluoromethoxy)-1-[(3aR,6aS)-hexahydrocyclopenta[c]pyrrol-2(1H)-yl]ethan-1-one; PHOSPHATE ION | PHOSPHATE ION; 2-(difluoromethoxy)-1-[(3aR,6aS)-hexahydrocyclopenta[c]pyrrol-2(1H)-yl]ethan-1-one | XDS | 99.90% | 4.6 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 19.7 | - | No | - | - | 8.9 | 60.2 | 44.6 | - | |||||
| 5sm0 | 2.09 Å | 2022-03-16 | X-ray | PO4, LQI | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 23.58% | 27.30% | 27.00% | PHOSPHATE ION; (1-benzofuran-2-yl)(4-methylpiperidin-1-yl)methanone | PHOSPHATE ION; (1-benzofuran-2-yl)(4-methylpiperidin-1-yl)methanone | XDS | 99.80% | 5.6 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 20.0 | - | No | - | - | 17.8 | 33.8 | 62.8 | - | |||||
| 5sm1 | 1.94 Å | 2022-03-16 | X-ray | PO4, WH1 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 22.92% | 26.04% | 24.00% | N-methyl-N-[2-(pyridin-2-yl)ethyl]benzamide; PHOSPHATE ION | PHOSPHATE ION; N-methyl-N-[2-(pyridin-2-yl)ethyl]benzamide | XDS | 99.90% | 6.2 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 19.7 | - | No | - | - | 26.2 | 13.0 | 74.4 | - | |||||
| 5sm2 | 1.78 Å | 2022-03-16 | X-ray | PO4, LQ3 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 25.01% | 26.97% | 18.90% | PHOSPHATE ION; (5S)-5-(difluoromethoxy)pyridin-2(5H)-one | PHOSPHATE ION; (5S)-5-(difluoromethoxy)pyridin-2(5H)-one | XDS | 99.80% | 6.6 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 17.6 | - | No | - | - | 19.8 | 18.7 | 70.5 | - | |||||
| 5sm3 | 2.20 Å | 2022-03-16 | X-ray | PO4, GT4 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 25.66% | 30.56% | 30.80% | ~{N}-(4-hydroxyphenyl)-2-methoxy-ethanamide; PHOSPHATE ION | PHOSPHATE ION; ~{N}-(4-hydroxyphenyl)-2-methoxy-ethanamide | XDS | 99.80% | 4.8 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 8.4 | - | No | - | - | 6.5 | 23.7 | 52.9 | - | |||||
| 5sm4 | 2.16 Å | 2022-03-16 | X-ray | AWD, PO4 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 22.85% | 27.05% | 27.50% | PHOSPHATE ION; ~{N}-(4-fluorophenyl)-4-methyl-piperazine-1-carboxamide | ~{N}-(4-fluorophenyl)-4-methyl-piperazine-1-carboxamide; PHOSPHATE ION | XDS | 100.00% | 5.6 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 26.6 | - | No | - | - | 19.2 | 48.0 | 61.5 | - | |||||
| 5sm5 | 1.95 Å | 2022-03-16 | X-ray | PO4, S5J | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 20.42% | 24.92% | 23.10% | 2-[4-(2-methoxyphenyl)piperazin-1-yl]ethanenitrile; PHOSPHATE ION | PHOSPHATE ION; 2-[4-(2-methoxyphenyl)piperazin-1-yl]ethanenitrile | XDS | 100.00% | 6.3 | Diamond (I04-1) | REFMAC | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 23.4 | - | No | - | - | 35.9 | 43.7 | 42.5 | - | |||||
| 5sm6 | 2.29 Å | 2022-03-16 | X-ray | K1A, PO4 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 22.90% | 27.77% | 28.50% | PHOSPHATE ION; 3-[(3,5-dimethyl-1,2-oxazol-4-yl)methyl]-5-methyl-1,3,4-thiadiazol-2(3H)-one | 3-[(3,5-dimethyl-1,2-oxazol-4-yl)methyl]-5-methyl-1,3,4-thiadiazol-2(3H)-one; PHOSPHATE ION | XDS | 100.00% | 4.9 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 20.7 | - | No | - | - | 15.3 | 47.8 | 53.0 | - | |||||
| 5sm7 | 1.95 Å | 2022-03-16 | X-ray | PO4, LPU | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 23.66% | 26.75% | 24.80% | 1-(methanesulfonyl)piperidin-4-ol; PHOSPHATE ION | PHOSPHATE ION; 1-(methanesulfonyl)piperidin-4-ol | XDS | 99.90% | 6.2 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 16.7 | - | No | - | - | 21.2 | 13.6 | 72.1 | - | |||||
| 5sm8 | 1.95 Å | 2022-03-16 | X-ray | WKA, PO4 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 22.39% | 26.72% | 24.40% | PHOSPHATE ION; N-(2,1,3-benzoxadiazol-4-yl)acetamide | N-(2,1,3-benzoxadiazol-4-yl)acetamide; PHOSPHATE ION | XDS | 100.00% | 5.3 | Diamond (I04-1) | REFMAC | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 16.0 | - | No | - | - | 21.4 | 40.0 | 43.7 | - | |||||
| 5sm9 | 2.01 Å | 2022-03-16 | X-ray | PO4, VZS | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 23.52% | 27.05% | 27.30% | PHOSPHATE ION; N-(2-methoxy-5-methylphenyl)-N'-4H-1,2,4-triazol-4-ylurea | PHOSPHATE ION; N-(2-methoxy-5-methylphenyl)-N'-4H-1,2,4-triazol-4-ylurea | XDS | 99.90% | 6.0 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 23.4 | - | No | - | - | 19.2 | 32.7 | 70.2 | - | |||||
| 5sma | 2.01 Å | 2022-03-16 | X-ray | NZD, PO4 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 22.67% | 25.91% | 24.70% | 4-methyl-N-phenylpiperazine-1-carboxamide; PHOSPHATE ION | 4-methyl-N-phenylpiperazine-1-carboxamide; PHOSPHATE ION | XDS | 100.00% | 6.0 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 27.0 | - | No | - | - | 27.2 | 34.1 | 68.2 | - | |||||
| 5smb | 2.18 Å | 2022-03-16 | X-ray | LRR, PO4 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 23.38% | 28.02% | 30.50% | PHOSPHATE ION; 1-(morpholin-4-yl)-4-phenylbutan-1-one | 1-(morpholin-4-yl)-4-phenylbutan-1-one; PHOSPHATE ION | XDS | 99.90% | 5.4 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 16.4 | - | No | - | - | 13.9 | 31.3 | 61.0 | - | |||||
| 5smc | 2.19 Å | 2022-03-16 | X-ray | PO4, LRF | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 20.85% | 26.31% | 26.10% | PHOSPHATE ION; N~2~-(4-cyano-3-methyl-1,2-thiazol-5-yl)-N~2~-methylglycinamide | PHOSPHATE ION; N~2~-(4-cyano-3-methyl-1,2-thiazol-5-yl)-N~2~-methylglycinamide | XDS | 99.50% | 6.4 | Diamond (I04-1) | REFMAC | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 23.8 | - | No | - | - | 24.3 | 48.0 | 50.6 | - | |||||
| 5smd | 1.83 Å | 2022-03-16 | X-ray | PO4, WKS | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 23.62% | 26.29% | 20.40% | PHOSPHATE ION; 2,4-dimethyl-6-(piperazin-1-yl)pyrimidine | PHOSPHATE ION; 2,4-dimethyl-6-(piperazin-1-yl)pyrimidine | XDS | 99.60% | 7.7 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 26.0 | - | No | - | - | 24.6 | 24.5 | 78.3 | - | |||||
| 5sme | 1.91 Å | 2022-03-16 | X-ray | I8D, PO4 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 21.26% | 25.53% | 22.20% | (4-chlorophenyl)(thiomorpholin-4-yl)methanone; PHOSPHATE ION | (4-chlorophenyl)(thiomorpholin-4-yl)methanone; PHOSPHATE ION | XDS | 99.90% | 6.6 | Diamond (I04-1) | REFMAC | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 23.7 | - | No | - | - | 30.5 | 36.9 | 55.2 | - | |||||
| 5smf | 2.01 Å | 2022-03-16 | X-ray | K1S, PO4 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 26.99% | 31.85% | 25.10% | N,N-diethyl-5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7-amine; PHOSPHATE ION | N,N-diethyl-5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7-amine; PHOSPHATE ION | XDS | 100.00% | 6.1 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 8.2 | - | No | - | - | 5.1 | 16.3 | 60.9 | - | |||||
| 5smg | 1.87 Å | 2022-03-16 | X-ray | PO4, LR9 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 23.13% | 25.31% | 19.00% | 3-amino-N-ethyl-N-methylbenzamide; PHOSPHATE ION | PHOSPHATE ION; 3-amino-N-ethyl-N-methylbenzamide | XDS | 99.90% | 7.1 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 25.1 | - | No | - | - | 32.4 | 23.0 | 70.3 | - | |||||
| 5smh | 2.64 Å | 2022-03-16 | X-ray | PO4, 7ZC | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 21.06% | 28.04% | 52.20% | PHOSPHATE ION; 1-(5-methoxy-1H-indol-3-yl)-N,N-dimethyl-methanamine | PHOSPHATE ION; 1-(5-methoxy-1H-indol-3-yl)-N,N-dimethyl-methanamine | XDS | 100.00% | 4.6 | Diamond (I04-1) | BUSTER | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 35.1 | - | No | - | - | 13.9 | 75.4 | 57.0 | - | |||||
| 5smi | 2.08 Å | 2022-03-16 | X-ray | PO4, LQV | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 26.82% | 32.90% | 32.50% | (2S)-N-(5-methylpyridin-2-yl)oxolane-2-carboxamide; PHOSPHATE ION | PHOSPHATE ION; (2S)-N-(5-methylpyridin-2-yl)oxolane-2-carboxamide | XDS | 99.80% | 5.4 | Diamond (I04-1) | REFMAC | 2022-03-03 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 4.3 | - | No | - | - | 4.5 | 26.2 | 36.2 | - | |||||
| 7mw2 | 2.97 Å | 2022-03-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-05-15 | 35347138 | - | - | - | - | N/A | - | - | Fab of antibody clone 6, light chain; Spike glycoprotein; Fab of antibody clone 6, heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7mw3 | 3.15 Å | 2022-03-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-05-15 | 35347138 | - | - | - | - | N/A | - | - | Fab of antibody clone 6, heavy chain; Spike glycoprotein; Fab of antibody clone 6, light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7mw4 | 3.42 Å | 2022-03-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-05-15 | 35347138 | - | - | - | - | N/A | - | - | Fab of antibody clone 6, light chain; Fab of antibody clone 6, heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7mw5 | 3.42 Å | 2022-03-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-05-15 | 35347138 | - | - | - | - | N/A | - | - | Fab of antibody clone 2, light chain; Spike glycoprotein; Fab of antibody clone 2, heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7mw6 | 3.22 Å | 2022-03-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-05-15 | 35347138 | - | - | - | - | N/A | - | - | Fab of antibody clone 2, light chain; Fab of antibody clone 2, heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7owx | 1.93 Å | 2022-03-16 | X-ray | - | SARS-CoV-2 | Spike | Protein-protein complex | 17.18% | 23.29% | N/A | Spike protein S2 | - | XDS | 99.30% | 11.5 | SOLEIL (PROXIMA 1) | PHENIX | 2021-06-21 | 35157347 | ZN | - | - | - | N/A | - | - | Spike protein S2 | 74.4 | - | No | - | 52.3 | 84.4 | 89.8 | - | ||||||
| 7qch | 1.88 Å | 2022-03-16 | X-ray | PO4, A5I | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 20.83% | 23.54% | N/A | PHOSPHATE ION; N-(3,5-dimetoxy-4-hydroxybenzyliden)thiosemicarbazone | PHOSPHATE ION; N-(3,5-dimetoxy-4-hydroxybenzyliden)thiosemicarbazone | DIALS | 99.85% | 12.94 | PETRA III, DESY (P11) | PHENIX | 2021-11-24 | 35480391 | ZN | - | - | - | N/A | - | - | Papain-like protease nsp3 | 30.7 | - | No | - | 49.6 | 14.7 | 73.1 | - | ||||||
| 7qci | 1.76 Å | 2022-03-16 | X-ray | PO4, A6Q | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 19.18% | 21.45% | N/A | PHOSPHATE ION; N-(3,4-dihydroxybenzylidene)-thiosemicarbazone | PHOSPHATE ION; N-(3,4-dihydroxybenzylidene)-thiosemicarbazone | DIALS | 99.91% | 13.32 | PETRA III, DESY (P11) | PHENIX | 2021-11-24 | 35480391 | ZN | - | - | - | N/A | - | - | Papain-like protease nsp3 | 48.7 | - | No | - | 70.2 | 16.7 | 85.7 | - | ||||||
| 7qcj | 1.84 Å | 2022-03-16 | X-ray | PO4, A4O | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 17.60% | 19.76% | N/A | N-(2,4-dihydroxybenzylidene)-thiosemicarbazone; PHOSPHATE ION | PHOSPHATE ION; N-(2,4-dihydroxybenzylidene)-thiosemicarbazone | DIALS | 99.95% | 10.95 | PETRA III, DESY (P11) | PHENIX | 2021-11-24 | 35480391 | ZN | - | - | - | N/A | - | - | Papain-like protease nsp3 | 56.3 | - | No | - | 83.3 | 17.1 | 86.7 | - | ||||||
| 7qck | 1.92 Å | 2022-03-16 | X-ray | A7L, PO4 | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 20.19% | 22.40% | N/A | PHOSPHATE ION; N-(2,5-dihydroxybenzylidene)-thiosemicarbazone | N-(2,5-dihydroxybenzylidene)-thiosemicarbazone; PHOSPHATE ION | DIALS | 99.88% | 9.33 | PETRA III, DESY (P11) | PHENIX | 2021-11-24 | 35480391 | ZN | - | - | - | N/A | - | - | Papain-like protease nsp3 | 39.2 | - | No | - | 61.1 | 13.7 | 79.5 | - | ||||||
| 7qcm | 1.77 Å | 2022-03-16 | X-ray | A3X, PO4 | SARS-CoV-2 | NSP3: PLpro | Functional ligand | 18.14% | 20.65% | N/A | N-(3-metoxy-4-hydroxy-acetophenone)thiosemicarbazone; PHOSPHATE ION | N-(3-metoxy-4-hydroxy-acetophenone)thiosemicarbazone; PHOSPHATE ION | DIALS | 99.95% | 14.0 | PETRA III, DESY (P11) | PHENIX | 2021-11-24 | 35480391 | ZN | - | - | - | N/A | - | - | Papain-like protease nsp3 | 54.5 | - | No | - | 76.9 | 27.3 | 79.4 | - | ||||||
| 7rnw | 2.35 Å | 2022-03-16 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.07% | 23.08% | 25.40% | ACE-DTY-LEU-GLN-TYR-ALA-VAL-LEU-ARG-HIS-LYS-ARG-ARG-GLU-SEC | - | XDS | 99.90% | 6.4 | Australian Synchrotron (MX2) | REFMAC | 2021-07-30 | 35432913 | - | - | - | - | N/A | - | - | 3C-like proteinase; ACE-DTY-LEU-GLN-TYR-ALA-VAL-LEU-ARG-HIS-LYS-ARG-ARG-GLU-SEC | 42.6 | - | No | - | 54.3 | 94.4 | 12.2 | - | ||||||
| 7tew | 3.52 Å | 2022-03-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-01-06 | 35136050 | - | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tey | 2.25 Å | 2022-03-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-01-06 | 35136050 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tez | 3.27 Å | 2022-03-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Pathogen-host interaction | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-01-06 | 35136050 | - | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7thm | 3.18 Å | 2022-03-16 | Cryo-EM | MN, POP | SARS-CoV-2 | NSP7/NSP8/NSP9/NSP12 | N/A | N/A | N/A | PYROPHOSPHATE 2-; MANGANESE (II) ION | MANGANESE (II) ION; PYROPHOSPHATE 2- | - | - | () | PHENIX | 2022-01-11 | 35194601 | MN; ZN | - | - | - | N/A | - | - | Non-structural protein 9; Non-structural protein 7; Non-structural protein 8; RNA-directed RNA polymerase - UNP residues 4393-5324 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wpd | 3.18 Å | 2022-03-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-01-23 | 35133176 | - | - | - | - | N/A | - | - | JMB2002 Fab heavy chain; Anti-Fab nanobody; JMB2002 Fab light chian; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wpf | 2.92 Å | 2022-03-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Protein-protein complex | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-23 | 35133176 | - | - | - | - | N/A | - | - | Spike glycoprotein; Anti-Fab nanobody; JMB2002 Fab light chain; JMB2002 Fab heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wue | 3.20 Å | 2022-03-16 | X-ray | - | SARS-CoV-2 | Spike | Protein-protein complex | 22.83% | 29.54% | 18.20% | - | iMOSFLM | 99.30% | 5.7 | NSRRC (BL15A1) | PHENIX | 2022-02-08 | 35230146 | - | - | - | - | N/A | - | - | m31A7 Fab HEAVY CHAIN; m31A7 Fab LIGHT CHAIN; Spike protein S1 | 11.7 | - | No | - | 8.4 | 63.3 | 22.1 | - | |||||||
| 5smk | 1.65 Å | 2022-03-23 | X-ray | PO4 | SARS-CoV-2 | NSP14 | PanDDA fragment screening | 19.06% | 21.73% | N/A | PHOSPHATE ION | PHOSPHATE ION | XDS | 91.50% | 9.1 | Diamond (I04-1) | REFMAC | 2022-03-08 | - | ZN | - | - | - | N/A | - | - | Proofreading exoribonuclease nsp14 | 46.8 | - | No | - | - | 67.7 | 31.3 | 69.9 | - | |||||
| 7e9p | 3.69 Å | 2022-03-23 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-03-04 | - | - | - | - | - | N/A | - | - | Heavy chain of 35B5 Fab; Spike glycoprotein; Light chain of 35B5 Fab | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7f46 | 4.79 Å | 2022-03-23 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-06-17 | - | - | - | - | - | N/A | - | - | Spike glycoprotein - UNP residues 1-1208; Heavy chain of 35B5 Fab; Light chain of 35B5 Fab | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7qcg | 1.75 Å | 2022-03-23 | X-ray | DZI, PO4 | SARS-CoV-2 | NSP3: PLpro | No functional ligands | 18.96% | 19.83% | N/A | PHOSPHATE ION; 3,4,5-tris(oxidanyl)-N-[(E)-1H-pyrrol-2-ylmethylideneamino]benzamide | 3,4,5-tris(oxidanyl)-N-[(E)-1H-pyrrol-2-ylmethylideneamino]benzamide; PHOSPHATE ION | DIALS | 99.96% | 12.39 | PETRA III, DESY (P11) | PHENIX | 2021-11-23 | 35480391 | ZN | - | - | - | N/A | - | - | Papain-like protease nsp3 | 58.5 | - | No | - | 82.8 | 20.9 | 87.8 | - | ||||||
| 7rby | 2.82 Å | 2022-03-23 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 20.06% | 28.68% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | iMOSFLM | 80.30% | 5.6 | SSRL (BL9-2) | PHENIX | 2021-07-06 | 35289719 | MG | - | - | - | N/A | - | - | Ilama-isolated nanobody NIH-CoV nb-112 specific to SARS-CoV-2 RBD; Spike protein S1 | 23.9 | - | No | - | 11.2 | 89.2 | 22.7 | - | ||||||
| 7tj2 | 3.20 Å | 2022-03-23 | Cryo-EM | - | SARS-CoV-2 | NSP15/RNA | Protein-protein complex | N/A | N/A | N/A | RNA (31-MER); RNA (31-MER) | - | - | - | () | PHENIX; phenix.real_space_refine | 2022-01-14 | 35801916 | - | - | - | - | N/A | - | - | RNA (31-MER); RNA (31-MER); Uridylate-specific endoribonuclease nsp15 | N/A | - | No | - | N/A | N/A | N/A | RNA (31-MER); RNA (31-MER) | - | ||||||
| 7tqv | 3.43 Å | 2022-03-23 | Cryo-EM | - | SARS-CoV-2 | NSP15/RNA | Protein-protein complex | N/A | N/A | N/A | RNA (33-MER); RNA (33-MER) | - | - | - | () | PHENIX; phenix.real_space_refine | 2022-01-27 | 35801916 | - | - | - | - | N/A | - | - | RNA (33-MER); Uridylate-specific endoribonuclease; RNA (33-MER) | N/A | - | No | - | N/A | N/A | N/A | RNA (33-MER); RNA (33-MER) | - | ||||||
| 7wpe | 2.69 Å | 2022-03-23 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-01-23 | 35133176 | - | - | - | - | N/A | - | - | Spike glycoprotein; JMB2002 Fab light chian; Anti-Fab nanobody; JMB2002 Fab heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wrv | 2.47 Å | 2022-03-23 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-01-27 | 35133176 | - | - | - | - | N/A | - | - | Spike glycoprotein; JMB2002 Fab light chain; JMB2002 Fab heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wuh | 4.70 Å | 2022-03-23 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-08 | 35230146 | - | - | - | - | N/A | - | - | m31A7 Fab light chain; m31A7 Fab heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7z1a | 2.59 Å | 2022-03-23 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 20.83% | 24.07% | 12.50% | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 100.00% | 14.6 | Diamond (I03) | REFMAC | 2022-02-24 | 35858383 | - | - | - | - | N/A | - | - | Spike protein S1; F2 Nanobody; H11 Nanobody | 19.9 | - | No | - | 44.3 | 15.0 | 55.0 | - | |||||||
| 7z1b | 2.30 Å | 2022-03-23 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 19.77% | 24.33% | 12.90% | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 100.00% | 12.0 | Diamond (I03) | REFMAC | 2022-02-24 | 35858383 | - | - | - | - | N/A | - | - | Spike protein S1; Nanobody F2; Nanobody A10 | 42.3 | - | No | - | 41.7 | 48.4 | 70.3 | - | |||||||
| 7z1c | 1.90 Å | 2022-03-23 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 17.49% | 19.94% | 4.30% | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 96.40% | 10.0 | Diamond (I03) | REFMAC | 2022-02-24 | 35858383 | - | - | - | - | N/A | - | - | Spike protein S1; Nanobody F2; Nanobody B5 | 53.3 | - | No | - | 82.1 | 14.2 | 85.2 | - | |||||||
| 7z1d | 1.55 Å | 2022-03-23 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 16.67% | 18.62% | 8.30% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 98.50% | 18.3 | Diamond (I03) | REFMAC | 2022-02-24 | 35858383 | - | - | - | - | N/A | - | - | Spike protein S1; H11-H6 nanobody | 72.3 | - | No | - | 89.6 | 39.3 | 92.1 | - | ||||||
| 7z1e | 1.59 Å | 2022-03-23 | X-ray | NO3, NAG | SARS-CoV-2 | Spike | Functional ligand | 16.10% | 20.23% | 4.60% | 2-acetamido-2-deoxy-beta-D-glucopyranose; NITRATE ION | NITRATE ION; 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 100.00% | 24.9 | Diamond (I03) | REFMAC | 2022-02-24 | 35858383 | - | - | - | - | N/A | - | - | Spike protein S1; H11-H4 Q98R H100E | 51.8 | - | No | - | 80.1 | 34.7 | 63.7 | - | ||||||
| 7s82 | 3.50 Å | 2022-03-30 | Cryo-EM | - | SARS-CoV-2 | NSP5 (3CLpro) | Protein-protein complex | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-09-17 | - | - | - | - | - | N/A | - | - | 3C-like proteinase - UNP residues 3259-3569 | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7tb8 | 2.83 Å | 2022-03-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-12-21 | 35324257 | - | - | - | - | N/A | - | - | The heavy chain of SARS-CoV-2 antiboddy antibody B1-182.1; The light chain of SARS-COV-2 antibody B1-182.1; Spike glycoprotein; The light chain of SARS-COV-2 antibody A19-61.1; The heavy chain of SARS-CoV-2 antibody A19-61.1 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tbf | 3.10 Å | 2022-03-30 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-12-21 | 35324257 | - | - | - | - | N/A | - | - | Heavy chain of SARS-CoV-2 antibody A19-61.1; Light chain of SARS-CoV-2 antibody B1-182.1; Spike glycoprotein; Light chain of SARS-CoV-2 antibody A19-61.1; Heavy chain of SARS-CoV-2 antibody B1-182.1 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7tc9 | 5.08 Å | 2022-03-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-12-23 | 35324257 | - | - | - | - | N/A | - | - | Light chain of antibody A19-46.1; Spike protein S1; Heavy chain of antibody A19-46.1 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tca | 3.85 Å | 2022-03-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-12-23 | 35324257 | - | - | - | - | N/A | - | - | Heavy chain of antibody A19-46.1; Spike glycoprotein; Light chain of antibody A19-46.1 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tcc | 3.86 Å | 2022-03-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-12-23 | 35324257 | - | - | - | - | N/A | - | - | Heavy chain of antibody A19-46.1; heavy chain of antibody B1-182.1; Light chain of antibody B1-182.1; Light chain of antibody A19-46.1; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7te1 | 3.50 Å | 2022-03-30 | X-ray | - | SARS-CoV-2 | Spike | PPI | 29.65% | 34.42% | 39.50% | - | XDS | 99.98% | 10.1 | APS (21-ID-E) | PHENIX | 2022-01-03 | 35303475 | - | - | - | - | N/A | - | - | Ab17 heavy chain; Spike protein S1; Ab17 light chain | 22.0 | - | No | - | 4.0 | 83.0 | 32.0 | - | |||||||
| 7tex | 3.27 Å | 2022-03-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-01-06 | 35136050 | - | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tf0 | 3.02 Å | 2022-03-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-01-06 | 35136050 | - | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tf1 | 3.57 Å | 2022-03-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-01-06 | 35136050 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tf2 | 3.62 Å | 2022-03-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-01-06 | 35136050 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tf3 | 2.25 Å | 2022-03-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-01-06 | 35136050 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tf4 | 3.01 Å | 2022-03-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-01-06 | 35136050 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tf5 | 3.16 Å | 2022-03-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-01-06 | 35136050 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7tqb | 3.10 Å | 2022-03-30 | X-ray | - | SARS-CoV-2 | Spike/RNA | PPI | 22.94% | 27.19% | N/A | RNA; DNA | - | xia2 | 98.70% | 3.78 | APS (22-ID) | PHENIX | 2022-01-26 | 35347133 | - | - | - | - | N/A | - | - | FAB S9.6 Heavy Chain; FAB S9.6 Light Chain; RNA; DNA | 24.5 | - | No | - | 18.5 | 85.1 | 20.8 | RNA | - | |||||
| 7u0d | 4.80 Å | 2022-03-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-17 | 35324257 | - | - | - | - | N/A | - | - | Light chain of SARS-CoV-2 antibody A19-46.1; Light chain of SARS-CoV-2 antibody B1-182.1; Heavy chain of SARS-CoV-2 antibody B1-182.1; Heavy chain of SARS-CoV-2 antibody A19-46.1; Surface glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7u0n | 2.61 Å | 2022-03-30 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 18.84% | 23.50% | 5.50% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 92.60% | 9.3 | SSRL (BL12-1) | PHENIX | 2022-02-18 | 35343765 | MG; ZN | - | - | - | N/A | - | - | Spike protein S1 - receptor binding domain; Angiotensin-converting enzyme 2 | 34.0 | - | No | - | 49.8 | 30.7 | 63.6 | - | ||||||
| 7vfa | 1.75 Å | 2022-03-30 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | PPI | 19.19% | 22.54% | N/A | - | HKL-3000 | 99.74% | 2.0 | SSRF (BL19U1) | PHENIX | 2021-09-11 | 35324337 | - | - | - | - | N/A | - | - | NB1A2; 3C-like proteinase | 38.4 | - | No | - | 59.8 | 15.9 | 77.1 | - | |||||||
| 7vfb | 2.00 Å | 2022-03-30 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | PPI | 23.55% | 26.36% | N/A | - | HKL-3000 | 96.45% | 2.0 | SSRF (BL19U1) | PHENIX | 2021-09-11 | 35324337 | - | - | - | - | N/A | - | - | 3C-like proteinase; nb2b4 | 11.6 | - | No | - | 24.1 | 19.6 | 49.7 | - | |||||||
| 7vic | 2.10 Å | 2022-03-30 | X-ray | ODN | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 21.59% | 24.22% | 12.90% | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one | CrysalisPro | 99.70% | 10.5 | SEALED TUBE () | REFMAC | 2021-09-26 | 35600064 | - | - | - | - | N/A | - | - | 3C-like proteinase | 19.9 | - | No | - | 42.7 | 19.3 | 52.2 | - | ||||||
| 7vtc | 2.54 Å | 2022-03-30 | X-ray | 4WI | MERS-CoV | NSP5 (3CLpro) | Functional ligand | 19.18% | 24.17% | 4.80% | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | XDS | 92.00% | 11.6 | SSRF (BL02U1) | PHENIX | 2021-10-28 | 35389231 | - | - | - | - | N/A | - | - | 3C-like proteinase | 46.7 | - | No | - | 43.4 | 86.0 | 39.5 | - | ||||||
| 7wph | 2.89 Å | 2022-03-30 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 22.86% | 26.94% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.81% | 8.31 | Australian Synchrotron (MX2) | PHENIX | 2022-01-23 | 36138032 | - | - | - | - | N/A | - | - | FAB light chain; Spike protein S1 - RBD; Fab heavy chain | 11.2 | - | No | - | 19.9 | 42.6 | 29.8 | - | ||||||
| 7e9n | 3.69 Å | 2022-04-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-03-04 | - | - | - | - | - | N/A | - | - | Heavy chain of 35B5 Fab; Light chain of 35B5 Fab; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7enf | 2.76 Å | 2022-04-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-04-16 | - | - | - | - | - | N/A | - | - | Heavy chain of Fab30; Spike glycoprotein - UNP residues 1-1208; Light chain of Fab30 | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7eng | 3.59 Å | 2022-04-06 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-04-16 | - | - | - | - | - | N/A | - | - | Heavy chain of Fab30; Spike glycoprotein - UNP residues 1-1208; Light chain of Fab30 | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7rzq | 2.09 Å | 2022-04-06 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | 2021-08-27 | 35363556 | - | - | - | - | N/A | - | - | Spike protein S2'; SARS-CoV-2 HR1 linked to a scaffold,Spike protein S2' | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7rzr | 2.27 Å | 2022-04-06 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | 2021-08-27 | 35363556 | - | - | - | - | N/A | - | - | SARS-CoV-2 HR1 D936Y linked to a scaffold,Spike protein S2'; Spike protein S2' | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7rzs | 2.52 Å | 2022-04-06 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | 2021-08-27 | 35363556 | - | - | - | - | N/A | - | - | SARS-CoV-2 HR1 L938F linked to a scaffold,Spike protein S2'; Spike protein S2' | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7rzt | 2.35 Å | 2022-04-06 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | 2021-08-27 | 35363556 | - | - | - | - | N/A | - | - | Spike protein S2'; SARS-CoV-2 HR1 S940F linked to a scaffold,Spike protein S2' | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7rzu | 2.30 Å | 2022-04-06 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | 2021-08-27 | 35363556 | - | - | - | - | N/A | - | - | SARS-CoV-2 HR1 A942S linked to a scaffold,Spike protein S2'; Spike protein S2' | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7rzv | 2.11 Å | 2022-04-06 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | 2021-08-27 | 35363556 | - | - | - | - | N/A | - | - | Spike protein S2'; SARS-CoV-2 HR1 linked to a scaffold,Spike protein S2' | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7smv | 1.93 Å | 2022-04-06 | X-ray | UED | FIPV | NSP5 (3CLpro) | Functional ligand | 19.08% | 24.49% | N/A | N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | XDS | 99.90% | 18.52 | CLSI (08B1-1) | PHENIX | 2021-10-26 | 35281564 | - | - | - | - | N/A | - | - | 3C-like proteinase | 15.3 | - | No | - | 40.4 | 29.7 | 33.4 | - | ||||||
| 7sna | 2.05 Å | 2022-04-06 | X-ray | UED | FIPV | NSP5 (3CLpro) | Functional ligand | 17.36% | 22.68% | N/A | N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | XDS | 99.74% | 15.94 | CLSI (08B1-1) | PHENIX | 2021-10-27 | 35281564 | - | - | - | - | N/A | - | - | 3C-like proteinase | 56.0 | - | No | - | 58.4 | 70.8 | 57.5 | - | ||||||
| 7tik | 2.40 Å | 2022-04-06 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | 2022-01-13 | 36940324 | - | - | - | - | N/A | - | - | Ferritin, Dps family protein and Spike protein S2' chimera; Spike protein S2' | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7vah | 1.49 Å | 2022-04-06 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | PPI | 19.32% | 21.66% | 3.70% | - | XDS | 98.80% | 21.62 | SSRF (BL19U1) | PHENIX | 2021-08-29 | 35380892 | - | - | - | - | N/A | - | - | 3C-like proteinase | 46.3 | - | No | - | 68.4 | 18.9 | 80.8 | - | |||||||
| 7vlo | 2.02 Å | 2022-04-06 | X-ray | 4WI | SARS-CoV | NSP5 (3CLpro) | Functional ligand | 20.38% | 23.44% | 2.60% | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | HKL-2000 | 95.70% | 9.8 | SSRF (BL17U1) | PHENIX; PHENIX | 2021-10-05 | 35389231 | - | - | - | - | N/A | - | - | 3C-like proteinase | 53.0 | - | No | - | 50.7 | 68.2 | 62.0 | - | ||||||
| 7vlp | 1.50 Å | 2022-04-06 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.46% | 21.91% | 3.50% | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | HKL-2000 | 97.70% | 15.0 | SSRF (BL17U1) | PHENIX | 2021-10-05 | 35389231 | - | - | - | - | N/A | - | - | Replicase polyprotein 1a | 57.7 | - | No | - | 65.8 | 39.4 | 84.8 | - | ||||||
| 7vlq | 1.94 Å | 2022-04-06 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.99% | 22.85% | 3.40% | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | HKL-2000 | 95.50% | 15.3 | SSRF (BL17U1) | PHENIX | 2021-10-05 | 35389231 | - | - | - | - | N/A | - | - | 3C-like proteinase | 68.5 | - | No | - | 56.6 | 70.5 | 85.2 | - | ||||||
| 7vth | 2.00 Å | 2022-04-06 | X-ray | 7XB | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.50% | 25.68% | 9.00% | 2-[4-[[4-[bis(fluoranyl)methoxy]-2-methyl-phenyl]amino]-2,6-bis(oxidanylidene)-3-[[3,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazin-1-yl]-N-methyl-ethanamide | 2-[4-[[4-[bis(fluoranyl)methoxy]-2-methyl-phenyl]amino]-2,6-bis(oxidanylidene)-3-[[3,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazin-1-yl]-N-methyl-ethanamide | CrysalisPro | 99.50% | 8.6 | ROTATING ANODE () | REFMAC | 2021-10-29 | 35352927 | - | - | - | - | N/A | - | - | 3C-like proteinase | 53.0 | - | No | - | 29.4 | 91.0 | 60.4 | - | ||||||
| 7vu6 | 1.80 Å | 2022-04-06 | X-ray | 7YY | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 22.41% | 27.90% | 6.00% | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | CrysalisPro | 99.80% | 20.5 | ROTATING ANODE () | REFMAC | 2021-11-01 | 35352927 | - | - | - | - | N/A | - | - | 3C-like proteinase | 25.7 | - | No | - | 14.5 | 73.1 | 39.4 | - | ||||||
| 7wvp | 3.70 Å | 2022-04-06 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | 2022-02-10 | 35228716 | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7wvq | 4.04 Å | 2022-04-06 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-02-10 | 35228716 | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ejy | 3.04 Å | 2022-04-13 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 21.44% | 25.35% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | 92.73% | 10.61 | SSRF (BL17U1) | PHENIX | 2021-04-03 | 34480123 | - | - | - | - | N/A | - | - | Spike protein S1 - Receptor binding domain; Light Chain of BD-503; Heavy Chain of BD-503 | 23.6 | - | No | - | 32.1 | 79.0 | 11.4 | - | |||||||
| 7ejz | 3.63 Å | 2022-04-13 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 24.11% | 28.41% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | 95.96% | 7.96 | SSRF (BL17U1) | PHENIX | 2021-04-03 | 34480123 | - | - | - | - | N/A | - | - | Light Chain of BD-503; Heavy Chain of BD-503; Spike protein S1 - Receptor binding domain | 14.7 | - | No | - | 12.2 | 81.6 | 8.1 | - | |||||||
| 7ek0 | 2.70 Å | 2022-04-13 | X-ray | - | SARS-CoV-2 | Spike | PPI | 21.39% | 26.60% | 23.20% | - | 94.68% | 16.64 | SSRF (BL17U1) | PHENIX | 2021-04-03 | 34480123 | - | - | - | - | N/A | - | - | Spike protein S1 - Receptor binding domain; Light Chain of BD-503; Heavy Chain of BD-503 | 17.6 | - | No | - | 22.2 | 56.6 | 30.2 | - | ||||||||
| 7fjo | 3.34 Å | 2022-04-13 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-08-04 | 35291264 | - | - | - | - | N/A | - | - | Spike glycoprotein; T6 light chain; T6 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7o05 | 1.94 Å | 2022-04-13 | X-ray | - | SARS-CoV-2 | Nucleocapsid | PPI | 17.72% | 23.30% | 16.70% | - | XDS | 96.20% | 6.2 | ALBA (XALOC) | REFMAC | 2021-03-25 | 35842466 | - | - | - | - | N/A | - | - | Nucleoprotein | 56.1 | - | No | - | 51.9 | 81.3 | 53.6 | - | |||||||
| 7o35 | 1.80 Å | 2022-04-13 | X-ray | GTP | SARS-CoV-2 | Nucleocapsid | Functional ligand | 15.58% | 20.33% | 6.60% | GUANOSINE-5'-TRIPHOSPHATE | GUANOSINE-5'-TRIPHOSPHATE | XDS | 99.40% | 17.8 | ALBA (XALOC) | REFMAC | 2021-04-01 | 35842466 | - | - | - | - | N/A | - | - | Nucleoprotein | 79.7 | - | No | - | 79.4 | 91.8 | 70.6 | - | ||||||
| 7o36 | 2.00 Å | 2022-04-13 | X-ray | GTP | SARS-CoV-2 | Nucleocapsid | Functional ligand | 18.98% | 24.06% | 8.20% | GUANOSINE-5'-TRIPHOSPHATE | GUANOSINE-5'-TRIPHOSPHATE | XDS | 95.50% | 8.9 | ALBA (XALOC) | REFMAC | 2021-04-01 | 35842466 | - | - | - | - | N/A | - | - | Nucleoprotein | 58.8 | - | No | - | 44.3 | 91.6 | 56.2 | - | ||||||
| 7t01 | 2.69 Å | 2022-04-13 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | 2021-11-29 | 34592170 | - | - | - | - | N/A | - | - | Spike protein S1; 54042-4 Fab - Heavy Chain; 54042-4 Fab - Light Chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7ult | 1.90 Å | 2022-04-13 | X-ray | FMT, NA | SARS-CoV-2 | NSP10/NSP16 | Functional ligand | 16.44% | 18.75% | 13.00% | FORMIC ACID; SODIUM ION | FORMIC ACID; SODIUM ION | HKL-3000 | 99.70% | 19.1 | APS (21-ID-D) | REFMAC | 2022-04-05 | - | ZN; NA | - | - | - | N/A | - | - | 2'-O-methyltransferase; Non-structural protein 10 | - | - | No | - | - | 89.1 | - | - | - | |||||
| 7wed | 3.50 Å | 2022-04-13 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-12-23 | 35090164 | - | - | - | - | N/A | - | - | Spike protein S1; The light chain of Fab XGv347; The heavy chain of Fab XGv347 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wlc | 4.00 Å | 2022-04-13 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-01-13 | 35090164 | - | - | - | - | N/A | - | - | Heavy chain of XGv282; Light chain of XGv282; Spike protein S1 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wo1 | 2.15 Å | 2022-04-13 | X-ray | 3XI | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.74% | 22.31% | N/A | N-[(2S)-3-methyl-1-[[(2S)-4-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]amino]pentan-2-yl]amino]-1-oxidanylidene-butan-2-yl]cyclohexanecarboxamide | N-[(2S)-3-methyl-1-[[(2S)-4-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]amino]pentan-2-yl]amino]-1-oxidanylidene-butan-2-yl]cyclohexanecarboxamide | 97.00% | 23.6 | SSRF (BL19U1) | PHENIX | 2022-01-20 | 35216507 | - | - | - | - | N/A | - | - | 3C-like proteinase | 53.9 | - | No | - | 62.0 | 33.2 | 87.3 | - | |||||||
| 7wo3 | 2.01 Å | 2022-04-13 | X-ray | 59S | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.67% | 24.30% | N/A | (2S)-2-[[(2S)-2-[[(E)-3-(4-methoxyphenyl)prop-2-enoyl]amino]-3-methyl-butanoyl]amino]-4-methyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]pentanamide | (2S)-2-[[(2S)-2-[[(E)-3-(4-methoxyphenyl)prop-2-enoyl]amino]-3-methyl-butanoyl]amino]-4-methyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]pentanamide | HKL-3000 | 98.30% | 10.4 | SSRF (BL19U1) | PHENIX | 2022-01-20 | 35216507 | - | - | - | - | N/A | - | - | 3C-like proteinase | 40.3 | - | No | - | 41.9 | 55.8 | 58.7 | - | ||||||
| 7wof | 1.72 Å | 2022-04-13 | X-ray | 5IZ | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.83% | 20.36% | N/A | (2S,3S)-3-methyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]-2-[[(E)-3-phenylprop-2-enoyl]amino]pentanamide | (2S,3S)-3-methyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]-2-[[(E)-3-phenylprop-2-enoyl]amino]pentanamide | HKL-3000 | 96.50% | 44.7 | SSRF (BL19U1) | PHENIX | 2022-01-21 | 35216507 | - | - | - | - | N/A | - | - | 3C-like proteinase | 58.6 | - | No | - | 79.2 | 21.8 | 90.7 | - | ||||||
| 7woh | 1.72 Å | 2022-04-13 | X-ray | 5IW | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.94% | 21.57% | N/A | (2S)-4-methyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]-2-[[(2S)-3-phenyl-2-[[(E)-3-phenylprop-2-enoyl]amino]propanoyl]amino]pentanamide | (2S)-4-methyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]-2-[[(2S)-3-phenyl-2-[[(E)-3-phenylprop-2-enoyl]amino]propanoyl]amino]pentanamide | 97.00% | 40.6 | SSRF (BL19U1) | PHENIX | 2022-01-21 | 35216507 | - | - | - | - | N/A | - | - | 3C-like proteinase | 42.1 | - | No | - | 69.2 | 18.1 | 72.8 | - | |||||||
| 7x7d | 2.92 Å | 2022-04-13 | X-ray | - | SARS-CoV-2 | Spike | PPI | 20.54% | 25.57% | N/A | - | 93.80% | 6.9 | SSRF (BL02U1) | PHENIX | 2022-03-09 | 35371009 | - | - | - | - | N/A | - | - | Spike protein S1 - Delta RBD; Nb22 | 14.5 | - | No | - | 30.3 | 20.8 | 50.4 | - | ||||||||
| 7z0x | 1.80 Å | 2022-04-13 | X-ray | EPE | SARS-CoV-2 | Spike | Functional ligand | 16.25% | 18.75% | 1.95% | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID | autoPROC | 92.00% | 20.3 | Diamond (I04) | PHENIX | 2022-02-23 | 35482816 | - | - | - | - | N/A | - | - | THSC20.HVTR26 Fab light chain; Spike protein S1; THSC20.HVTR26 Fab heavy chain | 68.7 | - | No | - | 89.1 | 30.7 | 93.0 | - | ||||||
| 7z0y | 2.95 Å | 2022-04-13 | X-ray | - | SARS-CoV-2 | Spike | PPI | 21.87% | 24.09% | 5.63% | - | autoPROC | 78.00% | 17.4 | Diamond (I04) | PHENIX | 2022-02-23 | 35482816 | - | - | - | - | N/A | - | - | Spike protein S1; THSC20.HVTR04 Fab heavy chain; THSC20.HVTR04 Fab light chain | 39.6 | - | No | - | 44.1 | 28.9 | 82.1 | - | |||||||
| 7pnm | 3.70 Å | 2022-04-20 | Cryo-EM | NAG | HC_OC43 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-09-07 | 35614127 | - | - | - | - | N/A | - | - | 46C12 antibody heavy chain; 46C12 antibody light chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7pnq | 3.70 Å | 2022-04-20 | Cryo-EM | NAG | HC_OC43 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-09-07 | 35614127 | - | - | - | - | N/A | - | - | Spike glycoprotein; 43E6 antibody light chain; 43E6 antibody heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7po5 | 3.90 Å | 2022-04-20 | Cryo-EM | NAG | HC_OC43 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-09-08 | 35614127 | - | - | - | - | N/A | - | - | Spike glycoprotein; 47C9 antibody light chain; 47C9 antibody heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7r40 | 2.90 Å | 2022-04-20 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-08 | 35471062 | - | - | - | - | N/A | - | - | 87G7 light chain variable region; 87G7 heavy chain variable region; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7rq6 | 4.18 Å | 2022-04-20 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-08-05 | 35123652 | - | - | - | - | N/A | - | - | Spike glycoprotein; CV3-13 Fab heavy chain; CV3-13 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7tpr | 2.39 Å | 2022-04-20 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-01-25 | 34751270 | - | - | - | - | N/A | - | - | Nanobody 8A2; Nanobody 7A3; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ub0 | 3.31 Å | 2022-04-20 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-03-14 | 35732171 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7ub5 | 3.35 Å | 2022-04-20 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-03-14 | 35732171 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7ub6 | 3.52 Å | 2022-04-20 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-03-14 | 35732171 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7x7e | 2.67 Å | 2022-04-20 | X-ray | PG4 | SARS-CoV-2 | Spike | PPI | 20.45% | 24.95% | N/A | TETRAETHYLENE GLYCOL | 99.80% | 12.3 | SSRF (BL18U1) | PHENIX | 2022-03-09 | 35371009 | - | - | - | - | N/A | - | - | Nb22; Spike protein S1 | 16.0 | - | No | - | 35.7 | 20.0 | 49.4 | - | ||||||||
| 7fjn | 3.25 Å | 2022-04-27 | Cryo-EM | NAG | HIV-1/SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-08-04 | 35291264 | - | - | - | - | N/A | - | - | Spike glycoprotein,Envelope glycoprotein; T6 heavy chain; T6 light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7fjs | 2.90 Å | 2022-04-27 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 21.71% | 28.49% | 22.70% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.70% | 3.4 | SSRF (BL18U1) | PHENIX | 2021-08-04 | 35291264 | - | - | - | - | N/A | - | - | Spike protein S1 - receptor binding domain; T6 heavy chain; T6 light chain | 7.9 | - | No | - | 12.0 | 28.5 | 40.9 | - | ||||||
| 7ql8 | 1.81 Å | 2022-04-27 | X-ray | NA, I70 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 23.45% | 30.02% | N/A | SODIUM ION; (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | SODIUM ION; (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | XDS | 86.10% | 3.57 | PETRA III, DESY (P11) | REFMAC | 2021-12-19 | 35807537 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 15.8 | - | No | - | 7.2 | 55.5 | 41.8 | - | ||||||
| 7sd5 | 1.53 Å | 2022-04-27 | X-ray | - | SARS-CoV-2 | Spike | PPI | 17.63% | 20.62% | 4.50% | - | XDS | 97.10% | 17.5 | APS (24-ID-E) | REFMAC | 2021-09-29 | 35438546 | - | - | - | - | N/A | - | - | 10-40 Heavy chain; 10-40 Light chain; Spike protein S1 - receptor binding domain (UNP residues 319-537) | 73.1 | - | No | - | 77.1 | 67.7 | 78.4 | - | |||||||
| 7si2 | 3.20 Å | 2022-04-27 | X-ray | - | SARS-CoV-2 | Spike | PPI | 19.84% | 26.12% | 18.00% | - | XDS | 100.00% | 7.5 | APS (24-ID-E) | PHENIX | 2021-10-12 | 35438546 | - | - | - | - | N/A | - | - | Spike protein S1 - receptor-binding domain (UNP reisdues 319-537); 10-28 Heavy Chain; 10-28 Light Chain | 38.7 | - | No | - | 25.7 | 84.7 | 43.1 | - | |||||||
| 7swn | 4.30 Å | 2022-04-27 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | 2021-11-20 | 35536154 | - | - | - | - | N/A | - | - | G32A4 Fab light chain; G32A4 Fab heavy chain; Spike protein S1 - RBD domain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7swo | 4.10 Å | 2022-04-27 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | 2021-11-20 | 35536154 | - | - | - | - | N/A | - | - | C98C7 Fab light chain; C98C7 Fab heavy chain; Spike protein S1 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7swp | 3.80 Å | 2022-04-27 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-11-20 | 35536154 | - | - | - | - | N/A | - | - | Spike protein S1; G32Q4 Fab heavy chain; G32Q4 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ttm | 2.24 Å | 2022-04-27 | X-ray | NAG | Bat-CoV-SHC014 | Spike | Functional ligand | 20.13% | 25.60% | 18.20% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 98.60% | 6.4 | APS (24-ID-C) | PHENIX | 2022-02-01 | 35438546 | - | - | - | - | N/A | - | - | 1040 light chain; 1040 heavy chain; Spike protein S1 - receptor-binding domain (UNP residues 307-528) | 41.8 | - | No | - | 29.8 | 48.6 | 80.9 | - | ||||||
| 7ttx | 2.80 Å | 2022-04-27 | X-ray | NAG | Bat-CoV-RaTG13 | Spike | Functional ligand | 22.22% | 32.11% | 17.30% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 98.90% | 8.8 | APS (24-ID-E) | PHENIX | 2022-02-02 | 35438546 | - | - | - | - | N/A | - | - | 1040 light chain; Spike glycoprotein - receptor-binding domain (UNP residues 319-541); 1040 heavy chain | 21.3 | - | No | - | 4.9 | 42.8 | 69.8 | - | ||||||
| 7uap | 2.80 Å | 2022-04-27 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-03-13 | 35447092 | - | - | - | - | N/A | - | - | C1520 Fab Light Chain; C1520 Fab Heavy Chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7uaq | 3.10 Å | 2022-04-27 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-03-13 | 35447092 | - | - | - | - | N/A | - | - | C1520 Fab Light Chain; Spike glycoprotein; C1520 Fab Heavy Chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7uar | 3.50 Å | 2022-04-27 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-03-13 | 35447092 | - | - | - | - | N/A | - | - | C1717 Fab Heavy Chain; Spike glycoprotein; C1717 Fab Light Chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7x7n | 4.47 Å | 2022-04-27 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | Synthetic peptide SIH-5; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-03-10 | 35654847 | - | - | - | - | N/A | - | - | Synthetic peptide SIH-5; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7z0p | 2.52 Å | 2022-04-27 | X-ray | I8H, NA | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.35% | 28.98% | 4.00% | (1~{R},2~{S},5~{S})-3-[(2~{S})-2-(~{tert}-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-6,6-dimethyl-~{N}-[(2~{S},3~{R})-4-(methylamino)-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]-3-azabicyclo[3.1.0]hexane-2-carboxamide; SODIUM ION | (1~{R},2~{S},5~{S})-3-[(2~{S})-2-(~{tert}-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-6,6-dimethyl-~{N}-[(2~{S},3~{R})-4-(methylamino)-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]-3-azabicyclo[3.1.0]hexane-2-carboxamide; SODIUM ION | XDS | 99.00% | 16.2 | PETRA III, DESY (P11) | REFMAC | 2022-02-23 | 35807537 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 13.6 | - | No | - | 10.0 | 64.3 | 24.8 | - | ||||||
| 7p2g | 2.50 Å | 2022-05-04 | X-ray | 4N0 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 22.44% | 27.74% | N/A | (4~{R})-~{N}-(4-iodophenyl)-2-oxidanylidene-3,4-dihydro-1~{H}-quinoline-4-carboxamide | (4~{R})-~{N}-(4-iodophenyl)-2-oxidanylidene-3,4-dihydro-1~{H}-quinoline-4-carboxamide | XDS | 98.65% | 8.38 | SLS (X06SA) | PHENIX | 2021-07-05 | 35169179 | - | - | - | - | N/A | - | - | 3C-like proteinase | 28.1 | - | No | - | 15.4 | 45.6 | 71.0 | - | ||||||
| 7qt5 | 2.26 Å | 2022-05-04 | X-ray | GWS | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.76% | 23.13% | 170.00% | 2-cyclohexyl-~{N}-pyridin-3-yl-ethanamide | 2-cyclohexyl-~{N}-pyridin-3-yl-ethanamide | xia2 | 98.60% | 9.9 | Diamond (I24) | REFMAC | 2022-01-14 | 35647922 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 61.1 | - | No | - | 53.7 | 95.7 | 47.2 | - | ||||||
| 7qt6 | 2.11 Å | 2022-05-04 | X-ray | RZJ | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.71% | 22.21% | 17.00% | 1-methyl-3,4-dihydro-2~{H}-quinoline-7-sulfonamide | 1-methyl-3,4-dihydro-2~{H}-quinoline-7-sulfonamide | xia2 | 99.70% | 7.9 | Diamond (I24) | REFMAC | 2022-01-14 | 35647922 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 68.0 | - | No | - | 63.0 | 88.6 | 59.8 | - | ||||||
| 7qt7 | 2.25 Å | 2022-05-04 | X-ray | UHV | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.29% | 21.63% | 16.20% | N-(5-tert-butyl-1,2-oxazol-3-yl)-N-[(1R)-2-[(4-methoxy-2-methylphenyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | N-(5-tert-butyl-1,2-oxazol-3-yl)-N-[(1R)-2-[(4-methoxy-2-methylphenyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | xia2 | 95.20% | 8.9 | Diamond (I24) | REFMAC | 2022-01-14 | 35647922 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 77.2 | - | No | - | 68.6 | 83.5 | 81.9 | - | ||||||
| 7qt8 | 2.01 Å | 2022-05-04 | X-ray | R8H | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.37% | 23.65% | 17.00% | (2~{R})-5-oxidanylidene-~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(pyridin-2-ylmethylamino)butan-2-yl]-1-(phenylmethyl)pyrrolidine-2-carboxamide | (2~{R})-5-oxidanylidene-~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(pyridin-2-ylmethylamino)butan-2-yl]-1-(phenylmethyl)pyrrolidine-2-carboxamide | xia2 | 95.20% | 9.9 | Diamond (I24) | REFMAC | 2022-01-14 | 35647922 | - | - | - | - | N/A | - | - | 3C-like proteinase | 56.9 | - | No | - | 48.4 | 81.2 | 58.7 | - | ||||||
| 7qt9 | 2.43 Å | 2022-05-04 | X-ray | UJ1 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 14.96% | 20.03% | 16.60% | N-(5-tert-butyl-1H-pyrazol-3-yl)-N-[(1R)-2-[(2-ethyl-6-methylphenyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | N-(5-tert-butyl-1H-pyrazol-3-yl)-N-[(1R)-2-[(2-ethyl-6-methylphenyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | xia2 | 97.90% | 10.8 | Diamond (I24) | REFMAC | 2022-01-14 | 35647922 | - | - | - | - | N/A | - | - | Non-structural protein 6 | 77.0 | - | No | - | 81.5 | 88.6 | 63.2 | - | ||||||
| 7rbu | 3.70 Å | 2022-05-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-07-06 | 35467422 | - | - | - | - | N/A | - | - | PVI.V6-14 Fab light chain, variable region; PVI.V6-14 Fab heavy chain, variable region; Spike protein S1 - N-terminal domain (UNP residues 14-307) | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7rbv | 3.60 Å | 2022-05-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-07-06 | 35467422 | - | - | - | - | N/A | - | - | PVI.V6-14 Fab light chain, variable region; PVI.V6-14 Fab heavy chain, variable region; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7sb3 | 3.30 Å | 2022-05-04 | Cryo-EM | 8Z9, NAG | HC_OC43 | Spike | Functional ligand | N/A | N/A | N/A | Sapienic acid; 2-acetamido-2-deoxy-beta-D-glucopyranose | Sapienic acid; 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-09-23 | 35507649 | - | - | - | - | N/A | - | - | Spike protein; Human polyclonal Fab model with polyalanine backbone - Heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7sb4 | 4.70 Å | 2022-05-04 | Cryo-EM | 8Z9, NAG | HC_OC43 | Spike | Functional ligand | N/A | N/A | N/A | Sapienic acid; 2-acetamido-2-deoxy-beta-D-glucopyranose | Sapienic acid; 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-09-23 | 35507649 | - | - | - | - | N/A | - | - | Spike protein; Human polyclonal Fab model with polyalanine backbone - Heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7sb5 | 4.20 Å | 2022-05-04 | Cryo-EM | NAG, PLM | HC_OC43 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; PALMITIC ACID | 2-acetamido-2-deoxy-beta-D-glucopyranose; PALMITIC ACID | - | - | () | 2021-09-23 | 35507649 | - | - | - | - | N/A | - | - | Spike protein; Human polyclonal Fab model with polyalanine backbone - Heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7sbv | 3.10 Å | 2022-05-04 | Cryo-EM | NAG, 8Z9 | HC_OC43 | Spike | Functional ligand | N/A | N/A | N/A | Sapienic acid; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose; Sapienic acid | - | - | () | 2021-09-26 | 35507649 | - | - | - | - | N/A | - | - | Spike protein; Human polyclonal Fab model with polyalanine backbone - Heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7sbw | 3.20 Å | 2022-05-04 | Cryo-EM | 8Z9, NAG | HC_OC43 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; Sapienic acid | Sapienic acid; 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-09-26 | 35507649 | - | - | - | - | N/A | - | - | Human polyclonal Fab model with polyalanine backbone - Heavy chain; Spike protein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7sbx | 3.00 Å | 2022-05-04 | Cryo-EM | NAG, 8Z9 | HC_OC43 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; Sapienic acid | 2-acetamido-2-deoxy-beta-D-glucopyranose; Sapienic acid | - | - | () | 2021-09-26 | 35507649 | - | - | - | - | N/A | - | - | Spike protein; Human polyclonal Fab model with polyalanine backbone - Heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7sby | 3.00 Å | 2022-05-04 | Cryo-EM | 8Z9, NAG | HC_OC43 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; Sapienic acid | Sapienic acid; 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-09-26 | 35507649 | - | - | - | - | N/A | - | - | Human polyclonal Fab model with polyalanine backbone - Heavy chain; Spike protein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7tgr | 1.68 Å | 2022-05-04 | X-ray | B1S, NA, K, K36 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.20% | 22.50% | 6.00% | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; SODIUM ION; POTASSIUM ION; (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | XDS | 97.80% | 11.2 | APS (24-ID-C) | PHENIX | 2022-01-09 | 35471084 | K; NA | - | - | - | N/A | - | - | 3C-like proteinase - UNP residues 3264-3567 | 68.7 | - | No | - | 60.1 | 55.9 | 96.9 | - | ||||||
| 7tia | 1.64 Å | 2022-05-04 | X-ray | XTP, SCN | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.26% | 19.05% | 5.50% | benzyl [(2S)-3-cyclopropyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamate; THIOCYANATE ION | benzyl [(2S)-3-cyclopropyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamate; THIOCYANATE ION | XDS | 98.10% | 15.5 | APS (24-ID-C) | PHENIX | 2022-01-13 | 35393402 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 69.2 | - | No | - | 87.5 | 42.4 | 84.0 | - | ||||||
| 7tiu | 1.65 Å | 2022-05-04 | X-ray | V46, PO4 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.81% | 19.57% | 4.30% | (1S,2S)-2-[(N-{[(3-chlorophenyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; PHOSPHATE ION | (1S,2S)-2-[(N-{[(3-chlorophenyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; PHOSPHATE ION | XDS | 98.20% | 17.6 | APS (24-ID-C) | PHENIX | 2022-01-14 | 35393402 | MG | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 56.3 | - | No | - | 84.6 | 30.8 | 71.8 | - | ||||||
| 7tiv | 2.08 Å | 2022-05-04 | X-ray | W48 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.58% | 21.57% | 25.60% | (1S,2S)-2-[(N-{[(3-chlorophenyl)methoxy]carbonyl}-3-cyclohexyl-L-alanyl)amino]-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid | (1S,2S)-2-[(N-{[(3-chlorophenyl)methoxy]carbonyl}-3-cyclohexyl-L-alanyl)amino]-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid | XDS | 98.50% | 9.0 | APS (24-ID-C) | PHENIX | 2022-01-14 | 35393402 | MG | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 40.7 | - | No | - | 69.2 | 17.4 | 70.6 | - | ||||||
| 7tiw | 1.68 Å | 2022-05-04 | X-ray | PO4, I54 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.39% | 19.85% | 5.90% | (1S,2S)-2-[(N-{[(2-chlorophenyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; PHOSPHATE ION | PHOSPHATE ION; (1S,2S)-2-[(N-{[(2-chlorophenyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | XDS | 98.50% | 14.4 | APS (24-ID-C) | PHENIX | 2022-01-14 | 35393402 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 52.0 | - | No | - | 82.7 | 29.2 | 66.9 | - | ||||||
| 7tix | 2.00 Å | 2022-05-04 | X-ray | Q56 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.22% | 21.49% | 14.50% | N~2~-{[(naphthalen-2-yl)methoxy]carbonyl}-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | N~2~-{[(naphthalen-2-yl)methoxy]carbonyl}-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | XDS | 98.10% | 10.6 | APS (24-ID-C) | PHENIX | 2022-01-14 | 35393402 | MG | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 42.0 | - | No | - | 70.0 | 30.9 | 58.9 | - | ||||||
| 7tiy | 1.79 Å | 2022-05-04 | X-ray | PO4, Y48 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.54% | 19.46% | 11.70% | PHOSPHATE ION; (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-[(N-{[(2,4,5-trifluorophenyl)methoxy]carbonyl}-L-leucyl)amino]propane-1-sulfonic acid | PHOSPHATE ION; (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-[(N-{[(2,4,5-trifluorophenyl)methoxy]carbonyl}-L-leucyl)amino]propane-1-sulfonic acid | XDS | 98.80% | 9.6 | APS (24-ID-C) | PHENIX | 2022-01-14 | 35393402 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 72.1 | - | No | - | 85.2 | 68.5 | 66.9 | - | ||||||
| 7tiz | 1.55 Å | 2022-05-04 | X-ray | SCN, NA, N63 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.04% | 19.06% | 5.60% | THIOCYANATE ION; (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[3-(trifluoromethyl)phenyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid; SODIUM ION | THIOCYANATE ION; SODIUM ION; (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[3-(trifluoromethyl)phenyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid | XDS | 97.80% | 16.7 | APS (24-ID-C) | PHENIX | 2022-01-14 | 35393402 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 51.3 | - | No | - | 87.5 | 20.9 | 69.2 | - | ||||||
| 7tj0 | 2.17 Å | 2022-05-04 | X-ray | S4L | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.61% | 21.73% | 18.20% | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | XDS | 98.10% | 7.3 | APS (24-ID-C) | PHENIX | 2022-01-14 | 35393402 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 72.4 | - | No | - | 67.7 | 83.4 | 70.2 | - | ||||||
| 7tty | 3.11 Å | 2022-05-04 | X-ray | - | Bat-CoV-WIV1 | Spike | PPI | 23.46% | 32.93% | 19.00% | - | XDS | 99.60% | 2.0 | APS (24-ID-E) | PHENIX; PHENIX | 2022-02-02 | 35438546 | - | - | - | - | N/A | - | - | 1040 light chain; Spike protein S1 - receptor-binding domain (UNP residues 322-515); 1040 heavy chain | 28.9 | - | No | - | 4.5 | 32.4 | 96.8 | - | |||||||
| 7u2d | 2.76 Å | 2022-05-04 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 21.95% | 25.86% | 11.40% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 97.60% | 6.4 | APS (23-ID-B) | PHENIX | 2022-02-23 | 35767670 | - | - | - | - | N/A | - | - | Spike protein S1 - Receptor binding domain, UNP residues 333-530; ADG20 light chain; ADG20 heavy chain | 18.8 | - | No | - | 27.8 | 41.0 | 43.1 | - | ||||||
| 7u2e | 2.85 Å | 2022-05-04 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 25.41% | 28.44% | 14.50% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 100.00% | 5.4 | SSRL (BL12-1) | PHENIX | 2022-02-23 | 35767670 | - | - | - | - | N/A | - | - | ADI-55688 light chain; Spike protein S1 - Receptor binding domain, UNP residues 333-530; ADI-55688 heavy chain | 8.6 | - | No | - | 12.1 | 38.4 | 33.4 | - | ||||||
| 7urq | 2.05 Å | 2022-05-04 | X-ray | NAG | SARS-CoV-2 | Spike | PPI | 17.74% | 20.79% | 12.80% | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-3000 | 93.30% | 19.9 | SSRL (BL12-2) | PHENIX | 2022-04-22 | 37334379 | - | - | - | - | N/A | - | - | COV11 Fab LIGHT CHAIN; Spike protein S1; COV11 Fab HEAVY CHAIN | 48.8 | - | No | - | 75.9 | 39.5 | 57.5 | - | |||||||
| 7urs | 2.40 Å | 2022-05-04 | X-ray | PO4, MPD, NAG | SARS-CoV-2 | Spike | Functional ligand | 17.85% | 22.36% | 12.60% | (4S)-2-METHYL-2,4-PENTANEDIOL; PHOSPHATE ION | PHOSPHATE ION; (4S)-2-METHYL-2,4-PENTANEDIOL; 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-3000 | 92.30% | 17.5 | SSRL (BL12-2) | PHENIX | 2022-04-22 | 37334379 | - | - | - | - | N/A | - | - | COV11 Fab HEAVY CHAIN; COV11 Fab HEAVY CHAIN; Spike protein S1 | 27.8 | - | No | - | 61.6 | 30.6 | 39.0 | - | ||||||
| 7we7 | 3.80 Å | 2022-05-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-12-23 | 35090164 | - | - | - | - | N/A | - | - | The light chain of Fab 282; Spike glycoprotein; Heavy chain of Fab 282 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7we8 | 3.50 Å | 2022-05-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-12-23 | 35090164 | - | - | - | - | N/A | - | - | Spike glycoprotein; Heavy chain of Fab 265; Light chain of Fab 265 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7we9 | 3.60 Å | 2022-05-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-12-23 | 35090164 | - | - | - | - | N/A | - | - | Spike glycoprotein; The heavy chain of Fab XGv289; The light chain of Fab XGv289 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7wea | 3.30 Å | 2022-05-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-12-23 | 35090164 | - | - | - | - | N/A | - | - | Spike glycoprotein; The heavy chain of Fab XGv347; The light chain of Fab XGv347 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7web | 3.70 Å | 2022-05-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-12-23 | 35090164 | - | - | - | - | N/A | - | - | Spike glycoprotein; The light chain of Fab XGv347; The heavy chain of Fab XGv347 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7wec | 3.30 Å | 2022-05-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-12-23 | 35090164 | - | - | - | - | N/A | - | - | Spike glycoprotein; The light chain of Fab XGv347; The heavy chain of Fab XGv347 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7wee | 4.00 Å | 2022-05-04 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-12-23 | 35090164 | - | - | - | - | N/A | - | - | Spike glycoprotein; The heavy chain of Fab XGv265; The light chain of Fab XGv265 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wef | 3.80 Å | 2022-05-04 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-12-23 | 35090164 | - | - | - | - | N/A | - | - | Spike protein S1; The heavy chain of Fab XGv289; The light chain of Fab XGv289 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7z3z | 3.10 Å | 2022-05-04 | Cryo-EM | STE, NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; STEARIC ACID | STEARIC ACID; 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-03-03 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7s6i | 3.20 Å | 2022-05-11 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-09-14 | 35549549 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7s6j | 3.40 Å | 2022-05-11 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-09-14 | 35549549 | - | - | - | - | N/A | - | - | J08 fragment antigen binding light chain variable domain; Spike glycoprotein; J08 fragment antigen binding heavy chain variable domain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7s6k | 3.40 Å | 2022-05-11 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-09-14 | 35549549 | - | - | - | - | N/A | - | - | Spike glycoprotein; J08 fragment antigen binding light chain variable domain; J08 fragment antigen binding heavy chain variable domain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7s6l | 4.00 Å | 2022-05-11 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-09-14 | 35549549 | - | - | - | - | N/A | - | - | Spike glycoprotein; J08 fragment antigen binding heavy chain variable domain; J08 fragment antigen binding light chain variable domain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7sbu | 2.53 Å | 2022-05-11 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 22.04% | 26.11% | 12.80% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.70% | 7.8 | SSRL (BL12-1) | PHENIX | 2021-09-25 | 35549549 | - | - | - | - | N/A | - | - | J08 Fab light chain; Spike protein S1 - Receptor binding domain, UNP residues 333-530; J08 Fab heavy chain | 28.7 | - | No | - | 25.7 | 51.2 | 56.2 | - | ||||||
| 7uv5 | 1.45 Å | 2022-05-11 | X-ray | - | SARS-CoV-2 | NSP3: PLpro | PPI | 13.64% | 17.86% | 7.50% | - | HKL-3000 | 99.40% | 8.0 | APS (19-ID) | REFMAC | 2022-04-29 | 37185902 | ZN | - | - | - | N/A | - | - | Ubiquitin; Ubiquitin; Papain-like protease nsp3 | 59.0 | - | No | - | 92.6 | 13.5 | 86.4 | - | |||||||
| 7whi | 2.93 Å | 2022-05-11 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-12-30 | 35344711 | - | - | - | - | N/A | - | - | Spike glycoprotein; Bn03_nano1; Bn03_nano2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7whj | 3.27 Å | 2022-05-11 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-12-30 | 35344711 | - | - | - | - | N/A | - | - | Bn03_nano1; Bn03_nano2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7whk | 3.01 Å | 2022-05-11 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-12-30 | 35344711 | - | - | - | - | N/A | - | - | Bn03_nano1; Spike glycoprotein; Bn03_nano2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7x9e | 2.60 Å | 2022-05-11 | X-ray | - | SARS-CoV-2 | Spike | Functional ligand | 19.49% | 24.32% | 13.40% | Spike peptide | - | HKL-3000 | 99.60% | 18.8 | SSRF (BL18U1) | PHENIX | 2022-03-15 | 35773398 | - | - | - | - | N/A | - | - | Spike peptide; 76E1 Fab Light Chain; 76E1 Fab Heavy Chain | 40.2 | - | No | - | 41.8 | 88.1 | 26.3 | - | ||||||
| 7epx | 3.00 Å | 2022-05-18 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-04-28 | 35443747 | - | - | - | - | N/A | - | - | Spike glycoprotein; heavy chain of GW01; light chain of GW01 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7fat | 1.99 Å | 2022-05-18 | X-ray | - | SARS-CoV-2 | Spike | PPI | 19.27% | 24.22% | N/A | - | HKL-3000 | 100.00% | 11.2 | SSRF (BL19U1) | PHENIX | 2021-07-07 | - | - | - | - | - | N/A | - | - | Spike protein S1 - Receptor-binding domain (RBD); nb_1A7 | 57.8 | - | No | - | - | 42.7 | 53.5 | 93.8 | - | ||||||
| 7qez | 2.89 Å | 2022-05-18 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 23.86% | 28.10% | 26.10% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.40% | 10.2 | SOLEIL (PROXIMA 2) | PHENIX | 2021-12-03 | 35704748 | - | - | - | - | N/A | - | - | CR3022 light chain; CR3022 heavy chain; CV2.1169 light chain; CV2.1169 IgA heavy chain; Spike protein S1 | 24.0 | - | No | - | 13.5 | 43.8 | 66.0 | - | ||||||
| 7qf0 | 2.30 Å | 2022-05-18 | X-ray | - | SARS-CoV-2 | Spike | Functional ligand | 18.49% | 22.13% | 14.70% | SODIUM ION | SODIUM ION | XDS | 99.90% | 17.4 | SOLEIL (PROXIMA 2) | PHENIX | 2021-12-03 | 35704748 | - | - | - | - | N/A | - | - | CV2.2325 heavy chain; CV2.2325 light chain; Spike protein S1 | 57.4 | - | No | - | 63.8 | 42.2 | 83.2 | - | ||||||
| 7qf1 | 2.80 Å | 2022-05-18 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 22.86% | 25.40% | 44.50% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 100.00% | 10.4 | SOLEIL (PROXIMA 2) | PHENIX | 2021-12-03 | 35704748 | - | - | - | - | N/A | - | - | Spike protein S1; CV2.6264 light chain; CV2.6264 heavy chain | 45.6 | - | No | - | 31.6 | 58.8 | 76.2 | - | ||||||
| 7wg6 | 3.40 Å | 2022-05-18 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-12-28 | 35120603 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7zdq | 3.20 Å | 2022-05-18 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-03-29 | 35849637 | - | - | - | - | N/A | - | - | Spike protein S1; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7f5g | 1.75 Å | 2022-05-25 | X-ray | - | SARS-CoV-2 | Spike | Functional ligand | 18.79% | 21.72% | 11.20% | alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | - | HKL-2000 | 100.00% | 14.0 | SSRF (BL18U1) | PHENIX | 2021-06-22 | 35460753 | - | - | - | - | N/A | - | - | Nanobody DL4; Spike glycoprotein - Receptor binding domain (RBD) | 39.3 | - | No | - | 67.7 | 21.8 | 64.9 | - | ||||||
| 7mxp | 4.46 Å | 2022-05-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-05-19 | 34898670 | - | - | - | - | N/A | - | - | LP5 Fab Heavy Chain; Spike glycoprotein; LP5 Fab Light Chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7qdg | 3.40 Å | 2022-05-25 | Cryo-EM | NAG, NDG | SARS-CoV-2 | Spike | Functional ligand | 40.74% | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-alpha-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-alpha-D-glucopyranose | - | - | () | REFMAC | 2021-11-27 | 35816514 | - | - | - | - | N/A | - | - | Spike glycoprotein,Fibritin | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7qdh | 4.20 Å | 2022-05-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | 55.59% | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | REFMAC | 2021-11-27 | 35816514 | - | - | - | - | N/A | - | - | Spike glycoprotein,Fibritin | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7wku | 2.60 Å | 2022-05-25 | X-ray | - | PDCoV | NSP5 (3CLpro) | Functional ligand | 19.05% | 24.10% | 17.20% | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | - | 99.60% | 2.6 | SSRF (BL19U1) | PHENIX | 2022-01-11 | 35336895 | - | - | - | - | N/A | - | - | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE; Peptidase C30 | 50.5 | - | No | - | 43.8 | 57.7 | 74.5 | - | |||||||
| 7wly | 3.40 Å | 2022-05-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-01-14 | 35436443 | - | - | - | - | N/A | - | - | Spike glycoprotein; Heavy chain of 35B5 Fab; Light chain of 35B5 Fab | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wlz | 2.98 Å | 2022-05-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-14 | 35436443 | - | - | - | - | N/A | - | - | Spike glycoprotein; Heavy chain of 35B5 Fab; Light chain of 35B5 Fab | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7x7o | 3.75 Å | 2022-05-25 | X-ray | - | SARS-CoV-2 | Spike | PPI | 30.33% | 33.58% | N/A | - | XDS | 99.90% | 7.1 | SPring-8 (BL32XU) | REFMAC; PHENIX | 2022-03-10 | 35543180 | - | - | - | - | N/A | - | - | UT28K Fab, light chain; UT28K Fab, heavy chain; Spike protein S1 | 1.9 | - | No | - | 4.2 | 28.2 | 20.2 | - | |||||||
| 7fau | 2.08 Å | 2022-06-01 | X-ray | - | SARS-CoV-2 | Spike | PPI | 19.84% | 22.82% | N/A | - | HKL-3000 | 100.00% | 15.0 | SSRF (BL19U1) | PHENIX | 2021-07-07 | 35276082 | ZN | - | - | - | N/A | - | - | NB_1B11; Spike protein S1 - Receptor-binding domain (RBD) | 53.7 | - | No | - | 56.8 | 35.0 | 90.5 | - | |||||||
| 7qur | 2.27 Å | 2022-06-01 | Cryo-EM | GE9, NAG, SIA, EIC | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2,3,5-tris(iodanyl)benzamide; N-acetyl-alpha-neuraminic acid; LINOLEIC ACID | 2,3,5-tris(iodanyl)benzamide; 2-acetamido-2-deoxy-beta-D-glucopyranose; N-acetyl-alpha-neuraminic acid; LINOLEIC ACID | - | - | () | PHENIX; phenix.real_space_refine | 2022-01-18 | 35737812 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7whh | 2.60 Å | 2022-06-01 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 19.16% | 23.05% | 26.00% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.77% | 11.3 | SSRF (BL02U1) | PHENIX | 2021-12-30 | - | ZN | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike glycoprotein - Omicron RBD | 31.8 | - | No | - | - | 54.5 | 34.8 | 50.3 | - | |||||
| 7ws0 | 3.20 Å | 2022-06-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-28 | 35477022 | - | - | - | - | N/A | - | - | Spike glycoprotein; 510A5 heavy chain; 510A5 light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7ws1 | 3.40 Å | 2022-06-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-28 | 35477022 | - | - | - | - | N/A | - | - | 510A5 heavy chain; Spike glycoprotein; 510A5 light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7ws2 | 3.30 Å | 2022-06-01 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-01-28 | 35477022 | - | - | - | - | N/A | - | - | Spike protein S1 - RBD; 510A5 light chain; 510A5 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ws3 | 3.60 Å | 2022-06-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-28 | 35477022 | - | - | - | - | N/A | - | - | 510A5 light chain; Spike glycoprotein; 510A5 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7ws4 | 3.70 Å | 2022-06-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-28 | 35477022 | - | - | - | - | N/A | - | - | 510A5 heavy chain; 510A5 light chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7ws5 | 3.70 Å | 2022-06-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-28 | 35477022 | - | - | - | - | N/A | - | - | 510A5 light chain; Spike glycoprotein; 510A5 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7ws6 | 3.80 Å | 2022-06-01 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-01-28 | 35477022 | - | - | - | - | N/A | - | - | 510A5 heavy chain; Spike protein S1 - RBD; 510A5 light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ws7 | 3.40 Å | 2022-06-01 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-01-28 | 35477022 | - | - | - | - | N/A | - | - | 510A5 heavy chain; Spike protein S1 - RBD; 510A5 light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ws8 | 3.00 Å | 2022-06-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-28 | 35477022 | - | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7ws9 | 2.90 Å | 2022-06-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-28 | 35477022 | - | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7wsa | 3.00 Å | 2022-06-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-28 | 35477022 | - | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike protein S1 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7wwl | 3.00 Å | 2022-06-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-02-13 | 35478188 | - | - | - | - | N/A | - | - | Spike glycoprotein; light chain of ZWD12; heavy chain of ZWD12 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7wwm | 2.80 Å | 2022-06-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-02-13 | 35478188 | - | - | - | - | N/A | - | - | Spike glycoprotein; heavy chain of ZWC6; heavy chain of ZWC6 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7zb6 | 2.12 Å | 2022-06-01 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | PPI | 26.08% | 29.77% | N/A | - | XDS | 91.90% | 7.6 | PETRA III, EMBL c/o DESY (P14 (MX2)) | PHENIX | 2022-03-23 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 6.0 | - | No | - | - | 7.8 | 11.8 | 54.2 | - | ||||||
| 7zb7 | 1.63 Å | 2022-06-01 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | PPI | 17.42% | 21.29% | N/A | - | XDS | 92.20% | 12.8 | PETRA III, EMBL c/o DESY (P14 (MX2)) | PHENIX | 2022-03-23 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 70.8 | - | No | - | - | 71.7 | 73.8 | 71.9 | - | ||||||
| 7zb8 | 2.48 Å | 2022-06-01 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | PPI | 24.64% | 33.19% | N/A | - | XDS | 83.90% | 6.7 | PETRA III, EMBL c/o DESY (P14 (MX2)) | PHENIX | 2022-03-23 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 7.1 | - | No | - | - | 4.3 | 15.4 | 58.9 | - | ||||||
| 7zf3 | 3.15 Å | 2022-06-01 | X-ray | IPA | SARS-CoV-2 | Spike | PPI | 20.76% | 26.61% | 49.10% | ISOPROPYL ALCOHOL | ISOPROPYL ALCOHOL | xia2 | 100.00% | 5.0 | Diamond (I03) | PHENIX | 2022-04-01 | 35662412 | - | - | - | - | N/A | - | - | Omi-3 light chain; EY6A light chain; Omi-3 heavy chain; Spike protein S1; EY6A heavy chain | 57.3 | - | No | - | 22.1 | 91.4 | 75.6 | - | ||||||
| 7zf5 | 5.32 Å | 2022-06-01 | X-ray | - | SARS-CoV-2 | Spike | PPI | 24.90% | 25.56% | 50.80% | - | xia2 | 100.00% | 4.6 | Diamond (I03) | PHENIX | 2022-04-01 | 35662412 | - | - | - | - | N/A | - | - | Beta-54 light chain; Beta-54 heavy chain; Spike protein S1; Omi-12 heavy chain; Omi-12 light chain | - | - | No | - | 30.3 | - | 32.6 | - | |||||||
| 7zf7 | 3.46 Å | 2022-06-01 | X-ray | NAG | SARS-CoV-2 | Spike | PPI | 25.84% | 26.90% | 70.30% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 99.60% | 3.1 | Diamond (I03) | PHENIX | 2022-04-01 | 35662412 | ZN | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike protein S1 | 14.0 | - | No | - | 20.1 | 12.5 | 67.6 | - | ||||||
| 7zf8 | 2.95 Å | 2022-06-01 | X-ray | - | SARS-CoV-2 | Spike | PPI | 22.01% | 25.84% | 49.00% | - | xia2 | 100.00% | 2.1 | Diamond (I03) | PHENIX | 2022-04-01 | 35662412 | - | - | - | - | N/A | - | - | COVOX-150 light chain; COVOX-150 heavy chain; Spike protein S1 | 38.4 | - | No | - | 27.9 | 53.2 | 71.6 | - | |||||||
| 7zf9 | 3.25 Å | 2022-06-01 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 22.15% | 26.48% | 62.90% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 99.90% | 3.2 | Diamond (I03) | PHENIX | 2022-04-01 | 35662412 | - | - | - | - | N/A | - | - | Spike protein S1; COVOX-150 heavy chain; COVOX-150 light chain | 32.7 | - | No | - | 23.2 | 39.7 | 78.5 | - | ||||||
| 7zfb | 3.08 Å | 2022-06-01 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 26.05% | 30.78% | 26.40% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 100.00% | 5.4 | Diamond (I04) | PHENIX | 2022-04-01 | 35662412 | - | - | - | - | N/A | - | - | Omi-31 heavy chain; Nanobody C1; Spike protein S1; Omi-31 light chain; Omi-18 heavy chain; Omi-18 light chain | 3.3 | - | No | - | 6.2 | 13.5 | 41.7 | - | ||||||
| 7zfc | 3.24 Å | 2022-06-01 | X-ray | - | SARS-CoV-2 | Spike | PPI | 24.42% | 29.87% | 26.50% | - | xia2 | 100.00% | 6.6 | Diamond (I04) | PHENIX | 2022-04-01 | 35662412 | - | - | - | - | N/A | - | - | Spike protein S1; Omi-31 heavy chain; Omi-31 light chain; Omi-18 heavy chain; Nanobody C1; Omi-18 light chain | 5.1 | - | No | - | 7.6 | 13.6 | 49.4 | - | |||||||
| 7zfd | 3.39 Å | 2022-06-01 | X-ray | - | SARS-CoV-2 | Spike | PPI | 28.56% | 33.77% | N/A | - | xia2 | 100.00% | 2.4 | Diamond (I03) | PHENIX | 2022-04-01 | 35662412 | - | - | - | - | N/A | - | - | Omi-25 heavy chain; Omi-25 light chain; Spike protein S1 | - | - | No | - | 4.1 | - | 54.8 | - | |||||||
| 7zfe | 3.25 Å | 2022-06-01 | X-ray | - | SARS-CoV-2 | Spike | PPI | 23.66% | 29.30% | 24.10% | - | xia2 | 87.80% | 6.6 | Diamond (I03) | PHENIX | 2022-04-01 | 35662412 | - | - | - | - | N/A | - | - | Omi-32 heavy chain; Nanobody C1; Spike protein S1; Omi-32 light chain | 15.6 | - | No | - | 9.0 | 44.9 | 50.2 | - | |||||||
| 7zr7 | 3.70 Å | 2022-06-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-05-03 | 35662412 | - | - | - | - | N/A | - | - | Omi-42 heavy chain; Spike glycoprotein,Fibritin; Omi-42 light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7zr8 | 3.70 Å | 2022-06-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-05-03 | 35662412 | - | - | - | - | N/A | - | - | Spike glycoprotein,Fibritin; Omi-38 Fab light chain; Omi-38 fab heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7zr9 | 4.00 Å | 2022-06-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-05-03 | 35662412 | - | - | - | - | N/A | - | - | Omi-2 Fab light chain; Omi-2 Fab heavy chain; Spike glycoprotein,Fibritin | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7zrc | 3.50 Å | 2022-06-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-05-04 | 35662412 | - | - | - | - | N/A | - | - | Omi-38 Fab Light Chain; Omi-38 Fab Heavy Chain; Spike glycoprotein,Fibritin | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7n45 | - | 2022-06-08 | NMR | - | HKU1-CoV | Nucleocapsid | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2021-06-03 | - | - | - | - | - | N/A | - | - | Nucleoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7qus | 2.39 Å | 2022-06-08 | Cryo-EM | NAG, EIC | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; LINOLEIC ACID | 2-acetamido-2-deoxy-beta-D-glucopyranose; LINOLEIC ACID | - | - | () | PHENIX; phenix.real_space_refine | 2022-01-18 | 35737812 | - | - | - | - | N/A | - | - | Spike glycoprotein,Fibritin | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7r4i | 3.20 Å | 2022-06-08 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-08 | 35547740 | - | - | - | - | N/A | - | - | Spike glycoprotein; Camel-derived nanobody 2.15 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7r4q | 3.60 Å | 2022-06-08 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-09 | 35547740 | - | - | - | - | N/A | - | - | Spike glycoprotein; Camel-derived nanobody 1.29 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7r4r | 3.90 Å | 2022-06-08 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-09 | 35547740 | - | - | - | - | N/A | - | - | Spike glycoprotein; Camel-derived nanobody 1.10 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7sd4 | - | 2022-06-08 | NMR | - | SARS-CoV-2 | Nucleocapsid | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2021-09-29 | 35638584 | - | - | - | - | N/A | - | - | Nucleoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7uhb | 3.00 Å | 2022-06-08 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | 2022-03-26 | 35412328 | - | - | - | - | N/A | - | - | Spike glycoprotein; Multivalent miniprotein inhibitor AHB2-2GS-SB175 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7uhc | 3.10 Å | 2022-06-08 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-03-26 | 35412328 | - | - | - | - | N/A | - | - | Spike glycoprotein; Multivalent miniprotein inhibitor AHB2-2GS-SB175 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7w6r | 2.60 Å | 2022-06-08 | X-ray | NAG | Bat-CoV-RaTG13 | Spike | Functional ligand | 19.23% | 19.35% | 17.10% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.90% | 10.5 | SSRF (BL19U1) | PHENIX; PHENIX | 2021-12-02 | - | ZN | - | - | - | N/A | - | - | Spike glycoprotein - RBD; Angiotensin-converting enzyme | 80.1 | - | No | - | - | 85.9 | 90.8 | 66.4 | - | |||||
| 7w6u | 2.56 Å | 2022-06-08 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 19.34% | 22.65% | 17.20% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 99.90% | 17.8 | SSRF (BL02U1) | PHENIX | 2021-12-02 | - | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme; Spike protein S1 | 34.6 | - | No | - | - | 58.7 | 25.5 | 61.1 | - | |||||
| 7wri | 3.03 Å | 2022-06-08 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-26 | 35821014 | ZN | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7wsk | 3.30 Å | 2022-06-08 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 22.26% | 27.52% | 45.30% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.70% | 5.9 | SSRF (BL10U2) | PHENIX | 2022-01-29 | 35821014 | ZN | - | - | - | N/A | - | - | Spike protein S1; Processed angiotensin-converting enzyme 2 | 7.9 | - | No | - | 16.5 | 31.0 | 34.0 | - | ||||||
| 7xby | 2.85 Å | 2022-06-08 | X-ray | NAG, BR | SARS-CoV-2 | Spike | Functional ligand | 22.82% | 26.26% | 16.00% | 2-acetamido-2-deoxy-beta-D-glucopyranose; BROMIDE ION | 2-acetamido-2-deoxy-beta-D-glucopyranose; BROMIDE ION | HKL-2000 | 99.70% | 13.1 | SSRF (BL02U1) | PHENIX | 2022-03-22 | 35729237 | ZN | - | - | - | N/A | - | - | Spike protein S1; Angiotensin-converting enzyme | 18.7 | - | No | - | 24.7 | 47.0 | 39.9 | - | ||||||
| 7zf4 | 4.18 Å | 2022-06-08 | X-ray | - | SARS-CoV-2 | Spike | PPI | 36.95% | 38.53% | 72.80% | - | xia2 | 100.00% | 2.5 | Diamond (I03) | PHENIX | 2022-04-01 | 35662412 | - | - | - | - | N/A | - | - | Spike protein S1; Nanobody F2; Omi-9 heavy chain; Omi-9 light chain | - | - | No | - | 3.5 | - | 31.7 | - | |||||||
| 7f0x | 2.80 Å | 2022-06-15 | X-ray | - | SARS-CoV-2 | Spike | PPI | 22.13% | 26.95% | N/A | - | XDS | 99.60% | 7.9 | SSRF (BL17U) | PHENIX | 2021-06-07 | - | - | - | - | - | N/A | - | - | Spike protein S1; Antibody | 34.9 | - | No | - | - | 19.9 | 83.8 | 42.1 | - | ||||||
| 7f12 | 3.15 Å | 2022-06-15 | X-ray | NAG | SARS-CoV-2 | Spike | PPI | 19.51% | 24.47% | 12.10% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.70% | 21.5 | SSRF (BL17U) | PHENIX | 2021-06-07 | - | - | - | - | - | N/A | - | - | Spike protein S1; Antibody | 51.9 | - | No | - | - | 40.5 | 75.9 | 62.4 | - | |||||
| 7f15 | 2.65 Å | 2022-06-15 | X-ray | NAG | SARS-CoV-2 | Spike | PPI | 20.10% | 24.55% | 11.30% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.90% | 25.8 | SSRF (BL17U) | PHENIX | 2021-06-07 | - | - | - | - | - | N/A | - | - | Spike protein S1; Antibody | 37.3 | - | No | - | - | 39.7 | 56.4 | 54.6 | - | |||||
| 7f7h | 3.19 Å | 2022-06-15 | X-ray | - | SARS-CoV-2 | Spike | PPI | 20.54% | 27.53% | N/A | - | HKL-2000 | 99.23% | 7.065 | APS (24-ID-E) | PHENIX | 2021-06-29 | - | - | - | - | - | N/A | - | - | Light chain of A8-1 Fab; Heavy chain of A8-1 Fab; Spike glycoprotein S1 - RBD domain | 18.4 | - | No | - | - | 16.5 | 69.7 | 24.6 | - | ||||||
| 7fcp | 2.40 Å | 2022-06-15 | X-ray | NAG, TRS | SARS-CoV-2 | Spike | PPI | 19.10% | 24.46% | 8.00% | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL | HKL-2000 | 100.00% | 22.4 | NFPSS (BL19U1) | REFMAC | 2021-07-15 | 35108220 | - | - | - | - | N/A | - | - | P14-44 antibody Fab fragment light chain; P5-22 antibody Fab fragment light chain; P5-22 antibody Fab fragment heavy chain; P14-44 antibody Fab fragment heavy chain; Spike protein S1 | 64.7 | - | No | - | 40.5 | 73.1 | 90.7 | - | ||||||
| 7fcq | 1.89 Å | 2022-06-15 | X-ray | NAG | SARS-CoV-2 | Spike | PPI | 16.47% | 20.06% | 7.80% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 100.00% | 23.9 | NFPSS (BL19U1) | REFMAC | 2021-07-15 | 35108220 | - | - | - | - | N/A | - | - | P14-44 antibody Fab fragment heavy chain; Spike protein S1; P14-44 antibody Fab fragment light chain | 60.9 | - | No | - | 81.3 | 27.9 | 87.1 | - | ||||||
| 7whc | 2.27 Å | 2022-06-15 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | PPI | 19.74% | 26.41% | N/A | - | HKL-2000 | 89.00% | 17.0 | PAL/PLS (7A (6B, 6C1)) | PHENIX | 2021-12-30 | 35563658 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 10.2 | - | No | - | 23.6 | 40.4 | 24.9 | - | |||||||
| 7xic | 3.30 Å | 2022-06-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-04-12 | 35607524 | - | - | - | - | N/A | - | - | heavy chain of STS165; light chain of STS165; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7xid | 3.30 Å | 2022-06-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-04-12 | 35607524 | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7xo4 | 3.24 Å | 2022-06-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-04-30 | 35641567 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7xo5 | 3.13 Å | 2022-06-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-05-01 | 35641567 | ZN | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7xo6 | 2.60 Å | 2022-06-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-05-01 | 35641567 | ZN | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7xo7 | 3.38 Å | 2022-06-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-05-01 | 35641567 | ZN | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7xo8 | 3.48 Å | 2022-06-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-05-01 | 35641567 | ZN | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7xo9 | 3.00 Å | 2022-06-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-05-01 | 35641567 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7xoa | 3.20 Å | 2022-06-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-05-01 | 35641567 | ZN | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7xob | 3.30 Å | 2022-06-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-05-01 | 35641567 | ZN | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7xoc | 3.00 Å | 2022-06-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-05-01 | 35641567 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7xod | 3.27 Å | 2022-06-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-05-01 | 35641567 | - | - | - | - | N/A | - | - | Heavy chain of JMB2002 Fab; Light chain of JMB2002 Fab; Spike glycoprotein; Nanobody | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7f3q | 3.30 Å | 2022-06-22 | Cryo-EM | NAG, MES | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | - | - | () | 2021-06-16 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; Light chain of A5-10 Fab; Heavy chain of A5-10 Fab; Light chain of A34-2 Fab; Heavy chain of A34-2 Fab | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7f5r | 3.01 Å | 2022-06-22 | X-ray | NAG | SARS-CoV-2 | Spike | PPI | 18.12% | 21.78% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 98.12% | 7.71 | SSRF (BL18U1) | PHENIX | 2021-06-22 | - | - | - | - | - | N/A | - | - | Spike protein S1; mink ACE2 | 54.2 | - | No | - | - | 67.3 | 88.3 | 27.5 | - | |||||
| 7fc5 | 2.89 Å | 2022-06-22 | X-ray | NAG | SARS-CoV-2 | Spike | PPI | 22.88% | 25.86% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 95.46% | 10.11 | SSRF (BL18U1) | PHENIX | 2021-07-13 | 35917815 | - | - | - | - | N/A | - | - | Spike protein S1; Angiotensin-converting enzyme | 6.8 | - | No | - | 27.8 | 17.7 | 31.9 | - | ||||||
| 7mb4 | 1.83 Å | 2022-06-22 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.71% | 22.01% | 2.80% | - | 98.70% | 12.5 | NSLS-II (17-ID-2) | PHENIX | 2021-03-31 | 35729165 | - | - | - | - | N/A | - | - | THR-SER-ALA-VAL-LEU-GLN; 3C-like proteinase | 68.4 | - | No | - | 64.9 | 66.0 | 81.3 | - | ||||||||
| 7mb5 | 1.60 Å | 2022-06-22 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.43% | 18.38% | 2.70% | - | 96.70% | 12.7 | NSLS-II (17-ID-2) | PHENIX | 2021-03-31 | 35729165 | - | - | - | - | N/A | - | - | SER-GLY-VAL-THR-PHE-GLN; 3C-like proteinase | 67.6 | - | No | - | 90.7 | 35.8 | 84.0 | - | ||||||||
| 7mb6 | 2.21 Å | 2022-06-22 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 22.28% | 27.23% | 3.30% | - | 99.40% | 12.7 | NSLS-II (17-ID-2) | PHENIX | 2021-03-31 | 35729165 | - | - | - | - | N/A | - | - | 3C-like proteinase; LYS-VAL-ALA-THR-VAL-GLN | 15.4 | - | No | - | 18.2 | 22.7 | 62.6 | - | ||||||||
| 7mb7 | 2.02 Å | 2022-06-22 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.01% | 22.41% | 3.30% | - | 98.30% | 10.2 | NSLS-II (17-ID-2) | PHENIX | 2021-03-31 | 35729165 | - | - | - | - | N/A | - | - | ASN-ARG-ALA-THR-LEU-GLN; 3C-like proteinase | 81.7 | - | No | - | 61.0 | 88.7 | 99.1 | - | ||||||||
| 7mb8 | 1.62 Å | 2022-06-22 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.85% | 19.49% | 3.50% | - | 98.70% | 11.7 | NSLS-II (17-ID-2) | PHENIX | 2021-03-31 | 35729165 | - | - | - | - | N/A | - | - | SER-ALA-VAL-LYS-LEU-GLN; 3C-like proteinase | 75.4 | - | No | - | 85.1 | 51.1 | 92.8 | - | ||||||||
| 7mb9 | 1.81 Å | 2022-06-22 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.89% | 22.76% | 3.50% | - | 98.10% | 10.1 | NSLS-II (17-ID-2) | PHENIX | 2021-03-31 | 35729165 | - | - | - | - | N/A | - | - | ARG-GLU-PRO-MET-LEU-GLN; 3C-like proteinase | 64.9 | - | No | - | 57.5 | 74.0 | 73.1 | - | ||||||||
| 7t70 | 2.35 Å | 2022-06-22 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.15% | 21.99% | 12.84% | - | HKL-2000 | 98.74% | 12.96 | ROTATING ANODE () | PHENIX | 2021-12-14 | 35729165 | - | - | - | - | N/A | - | - | 3C-like proteinase; Nonstructural protein 4/5 | 44.6 | - | No | - | 65.3 | 35.3 | 64.0 | - | |||||||
| 7t8m | 1.60 Å | 2022-06-22 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | PPI | 16.05% | 19.71% | 5.00% | - | CrysalisPro | 92.38% | 22.08 | ROTATING ANODE () | PHENIX | 2021-12-16 | 35729165 | - | - | - | - | N/A | - | - | Nonstructural protein 5/6; 3C-like proteinase | 65.9 | - | No | - | 83.6 | 49.9 | 73.4 | - | |||||||
| 7t8r | 1.74 Å | 2022-06-22 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | PPI | 18.45% | 22.78% | 6.85% | - | CrysalisPro | 98.53% | 22.39 | ROTATING ANODE () | PHENIX | 2021-12-16 | 35729165 | - | - | - | - | N/A | - | - | 3C-like proteinase; Nonstructural protein 7/8 | 32.2 | - | No | - | 57.4 | 43.4 | 39.7 | - | |||||||
| 7t9y | 2.18 Å | 2022-06-22 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | PPI | 17.94% | 23.63% | 8.74% | Nonstructural protein 8/9 | - | CrysalisPro | 98.24% | 14.33 | ROTATING ANODE () | PHENIX | 2021-12-20 | 35729165 | - | - | - | - | N/A | - | - | Nonstructural protein 8/9; 3C-like proteinase | 52.4 | - | No | - | 48.6 | 49.6 | 81.6 | - | ||||||
| 7ta4 | 1.78 Å | 2022-06-22 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | PPI | 17.50% | 21.63% | 5.30% | Nonstructural protein 9/10 | - | CrysalisPro | 98.48% | 9.46 | ROTATING ANODE () | PHENIX | 2021-12-20 | 35729165 | - | - | - | - | N/A | - | - | Nonstructural protein 9/10; 3C-like proteinase | 57.5 | - | No | - | 68.6 | 45.1 | 75.8 | - | ||||||
| 7ta7 | 2.28 Å | 2022-06-22 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | PPI | 20.48% | 25.45% | 9.10% | Nonstructural protein 10/11 | - | CrysalisPro | 99.41% | 9.62 | ROTATING ANODE () | PHENIX | 2021-12-20 | 35729165 | - | - | - | - | N/A | - | - | Nonstructural protein 10/11; 3C-like proteinase | 20.7 | - | No | - | 31.2 | 21.8 | 63.1 | - | ||||||
| 7tb2 | 1.80 Å | 2022-06-22 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | PPI | 18.76% | 21.79% | 2.33% | Nonstructural protein 12/13 | - | CrysalisPro | 98.90% | 19.45 | ROTATING ANODE () | PHENIX | 2021-12-21 | 35729165 | - | - | - | - | N/A | - | - | Nonstructural protein 12/13; 3C-like proteinase | 54.7 | - | No | - | 67.2 | 38.3 | 78.6 | - | ||||||
| 7tbt | 2.45 Å | 2022-06-22 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | PPI | 21.02% | 26.64% | 6.68% | Nonstructural protein 13/14 | - | CrysalisPro | 95.40% | 9.58 | ROTATING ANODE () | PHENIX | 2021-12-22 | 35729165 | - | - | - | - | N/A | - | - | 3C-like proteinase; Nonstructural protein 13/14 | 26.4 | - | No | - | 22.0 | 50.0 | 56.3 | - | ||||||
| 7tc4 | 1.94 Å | 2022-06-22 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | PPI | 18.21% | 22.67% | 9.07% | Nonstructural protein 15/16 | - | CrysalisPro | 99.83% | 12.74 | ROTATING ANODE () | PHENIX | 2021-12-22 | 35729165 | - | - | - | - | N/A | - | - | Nonstructural protein 15/16; 3C-like proteinase | 37.1 | - | No | - | 58.5 | 34.1 | 57.6 | - | ||||||
| 7tq2 | 2.30 Å | 2022-06-22 | X-ray | ISG | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.10% | 26.29% | 11.20% | N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[(1R,2R)-2-phenylcyclopropyl]methoxy}carbonyl)-L-leucinamide | N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[(1R,2R)-2-phenylcyclopropyl]methoxy}carbonyl)-L-leucinamide | XDS | 99.90% | 7.3 | APS (17-ID) | PHENIX | 2022-01-26 | 36654747 | - | - | - | - | N/A | - | - | 3C-like proteinase | 54.1 | - | No | - | 24.6 | 88.4 | 70.0 | - | ||||||
| 7tq3 | 2.00 Å | 2022-06-22 | X-ray | IS5 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.23% | 24.68% | 5.90% | N~2~-({[(1R,2R)-2-(3-fluorophenyl)cyclopropyl]methoxy}carbonyl)-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | N~2~-({[(1R,2R)-2-(3-fluorophenyl)cyclopropyl]methoxy}carbonyl)-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | XDS | 98.90% | 12.2 | APS (17-ID) | PHENIX | 2022-01-26 | 36654747 | - | - | - | - | N/A | - | - | 3C-like proteinase | 61.6 | - | No | - | 38.5 | 88.3 | 71.1 | - | ||||||
| 7tq4 | 2.45 Å | 2022-06-22 | X-ray | IRZ | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 21.27% | 29.63% | 9.80% | N~2~-({[(1R,2R)-2-(3-chlorophenyl)cyclopropyl]methoxy}carbonyl)-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | N~2~-({[(1R,2R)-2-(3-chlorophenyl)cyclopropyl]methoxy}carbonyl)-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | XDS | 99.70% | 8.5 | APS (17-ID) | PHENIX | 2022-01-26 | 36654747 | - | - | - | - | N/A | - | - | 3C-like proteinase | 34.2 | - | No | - | 8.1 | 58.6 | 77.6 | - | ||||||
| 7uxx | 1.85 Å | 2022-06-22 | X-ray | - | SARS-CoV-2 | Nucleocapsid | No functional ligands | 16.28% | 20.70% | N/A | - | XDS | 99.95% | 9.33 | LNLS SIRUS (MANACA) | REFMAC | 2022-05-06 | 36323732 | - | - | - | - | N/A | - | - | Nucleoprotein | 78.1 | - | No | - | 76.5 | 95.2 | 65.0 | - | |||||||
| 7uxz | 1.73 Å | 2022-06-22 | X-ray | GKP, NA | SARS-CoV-2 | Nucleocapsid | Functional ligand | 17.40% | 21.49% | N/A | (2R,3R)-2,3-bis{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}butanedioic acid; SODIUM ION | (2R,3R)-2,3-bis{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}butanedioic acid; SODIUM ION | autoPROC | 99.86% | 9.46 | LNLS SIRUS (MANACA) | REFMAC | 2022-05-06 | 36323732 | - | - | - | - | N/A | - | - | Nucleoprotein | 65.1 | - | No | - | 70.0 | 82.3 | 52.8 | - | ||||||
| 7vrv | 4.20 Å | 2022-06-22 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-10-25 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7vrw | 3.60 Å | 2022-06-22 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-10-25 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||
| 7wg7 | 4.00 Å | 2022-06-22 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-12-28 | 35120603 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7wg8 | 3.90 Å | 2022-06-22 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-12-28 | 35120603 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7wg9 | 3.50 Å | 2022-06-22 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-12-28 | 35120603 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7wgb | 3.50 Å | 2022-06-22 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-12-28 | 35120603 | - | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7wgc | 3.60 Å | 2022-06-22 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-12-28 | 35120603 | - | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7whz | 3.42 Å | 2022-06-22 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-01 | 35594869 | - | - | - | - | N/A | - | - | IG c934_light_IGKV1-5_IGKJ1; IG c1437_light_IGLV1-40_IGLJ1; IGL c4029_light_IGKV1-39_IGKJ2; XMA01 heavy chain variable domain; XMA04 heavy chain variable domain; Spike protein S1; XMA09 heavy chain variable domain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7wi0 | 3.82 Å | 2022-06-22 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-01 | 35594869 | - | - | - | - | N/A | - | - | XMA09 heavy chain variable domain; IG c934_light_IGKV1-5_IGKJ1; IGL c4029_light_IGKV1-39_IGKJ2; XMA01 heavy chain variable domain; XMA04 heavy chain variable domain; Spike protein S1; IG c1437_light_IGLV1-40_IGLJ1 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7wr8 | 3.50 Å | 2022-06-22 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-01-26 | 35714668 | - | - | - | - | N/A | - | - | BD55-3152L; BD55-3152H; Spike protein S1 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wrl | 3.51 Å | 2022-06-22 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-01-27 | 35714668 | - | - | - | - | N/A | - | - | BD55-1239H; Spike protein S1; BD55-1239L | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wro | 3.40 Å | 2022-06-22 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-01-27 | 35714668 | - | - | - | - | N/A | - | - | 3372L; Spike protein S1 - RBD; 3372H | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wrz | 3.26 Å | 2022-06-22 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-01-28 | 35714668 | - | - | - | - | N/A | - | - | Spike protein S1; BD55-5840H; BD55-5840L | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wym | 2.05 Å | 2022-06-22 | X-ray | G7L | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.40% | 22.07% | N/A | N-methyl-N-[[4-(trifluoromethyl)-1,3-thiazol-2-yl]methyl]prop-2-enamide | N-methyl-N-[[4-(trifluoromethyl)-1,3-thiazol-2-yl]methyl]prop-2-enamide | XDS | 96.30% | 12.9 | SSRF (BL19U1) | PHENIX | 2022-02-16 | 35702321 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 63.8 | - | No | - | 64.5 | 78.7 | 59.2 | - | ||||||
| 7wyp | 2.30 Å | 2022-06-22 | X-ray | G7O | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 22.38% | 27.26% | N/A | N-(1,3-benzothiazol-2-ylmethyl)-N-cyclopropyl-prop-2-enamide | N-(1,3-benzothiazol-2-ylmethyl)-N-cyclopropyl-prop-2-enamide | XDS | 99.90% | 11.02 | SSRF (BL19U1) | PHENIX | 2022-02-16 | 35702321 | - | - | - | - | N/A | - | - | 3C-like proteinase | 13.8 | - | No | - | 18.1 | 30.1 | 51.3 | - | ||||||
| 8ctk | 3.52 Å | 2022-06-22 | Cryo-EM | - | SARS-CoV-2 | Membrane_glycoprotein | No functional ligands | N/A | N/A | N/A | - | - | - | () | phenix.real_space_refine; PHENIX | 2022-05-15 | 36264056 | - | - | - | - | N/A | - | - | Membrane protein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8czt | 2.10 Å | 2022-06-22 | X-ray | P8L | MERS-CoV | NSP5 (3CLpro) | Functional ligand | 20.04% | 26.17% | 6.60% | [(1~{S},2~{R})-2-cyclohexylcyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | [(1~{S},2~{R})-2-cyclohexylcyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | XDS | 96.50% | 10.6 | NSLS-II (19-ID) | PHENIX | 2022-05-25 | 36654747 | - | - | - | - | N/A | - | - | 3C-like proteinase | 66.6 | - | No | - | 25.4 | 95.7 | 87.2 | - | ||||||
| 8czu | 2.70 Å | 2022-06-22 | X-ray | P9R | MERS-CoV | NSP5 (3CLpro) | Functional ligand | 21.24% | 25.84% | 14.10% | [(1~{S},2~{S})-2-(cyclohexylmethyl)cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | [(1~{S},2~{S})-2-(cyclohexylmethyl)cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | XDS | 95.70% | 7.0 | NSLS-II (19-ID) | PHENIX | 2022-05-25 | 36654747 | - | - | - | - | N/A | - | - | 3C-like proteinase | 66.6 | - | No | - | 27.9 | 95.7 | 84.6 | - | ||||||
| 8czv | 1.95 Å | 2022-06-22 | X-ray | P8C, PJR | MERS-CoV | NSP5 (3CLpro) | Functional ligand | 18.41% | 24.17% | 8.90% | [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate; [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate; [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | XDS | 99.50% | 9.8 | NSLS-II (19-ID) | PHENIX | 2022-05-25 | 36654747 | - | - | - | - | N/A | - | - | 3C-like proteinase | 75.0 | - | No | - | 43.3 | 95.7 | 88.9 | - | ||||||
| 8czw | 1.70 Å | 2022-06-22 | X-ray | P8L, P8U, PG4 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.82% | 21.65% | 5.60% | [(1~{S},2~{R})-2-cyclohexylcyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate; [(1~{S},2~{R})-2-cyclohexylcyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | [(1~{S},2~{R})-2-cyclohexylcyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate; [(1~{S},2~{R})-2-cyclohexylcyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate; TETRAETHYLENE GLYCOL | XDS | 98.80% | 11.2 | NSLS-II (19-ID) | PHENIX | 2022-05-25 | 36654747 | - | - | - | - | N/A | - | - | 3C-like proteinase | 73.3 | - | No | - | 68.4 | 63.8 | 91.3 | - | ||||||
| 8czx | 1.65 Å | 2022-06-22 | X-ray | P8C, PG4, PJR | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.80% | 22.04% | 3.90% | [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate; [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate; TETRAETHYLENE GLYCOL; [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | XDS | 98.30% | 11.5 | NSLS-II (19-ID) | PHENIX | 2022-05-25 | 36654747 | - | - | - | - | N/A | - | - | 3C-like proteinase | 72.5 | - | No | - | 64.7 | 66.2 | 90.7 | - | ||||||
| 7f5h | 3.00 Å | 2022-06-29 | X-ray | PO4 | SARS-CoV-2 | Spike | PPI | 22.64% | 24.76% | 17.40% | PHOSPHATE ION | PHOSPHATE ION | HKL-2000 | 100.00% | 14.1 | SSRF (BL19U1) | REFMAC | 2021-06-22 | 35722331 | - | - | - | - | N/A | - | - | Nanobody DL28; SARS-CoV-2 Spike Receptor-Binding Domain (RBD) - Receptor binding domain (RBD) | 66.4 | - | No | - | 37.6 | 93.4 | 76.9 | - | ||||||
| 7f6y | 3.00 Å | 2022-06-29 | X-ray | NAG | SARS-CoV-2 | Spike | PPI | 19.75% | 25.30% | 20.00% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.49% | 8.36 | Photon Factory (BL-1A) | PHENIX | 2021-06-26 | 34480123 | - | - | - | - | N/A | - | - | Spike protein S1 - receptor binding domain; Light Chain of BD-503; Heavy Chain of BD-503 | 32.3 | - | No | - | 32.4 | 93.4 | 14.7 | - | ||||||
| 7f6z | 3.00 Å | 2022-06-29 | X-ray | - | SARS-CoV-2 | Spike | PPI | 20.68% | 25.40% | 20.00% | - | XDS | 99.45% | 8.33 | Photon Factory (BL-1A) | PHENIX | 2021-06-26 | 34480123 | - | - | - | - | N/A | - | - | Heavy Chain of BD-503; Spike protein S1 - receptor binding domain; Light Chain of BD-503 | 42.9 | - | No | - | 31.6 | 86.4 | 43.6 | - | |||||||
| 7r1t | 2.70 Å | 2022-06-29 | X-ray | 6NR, PO4, MES | SARS-CoV-2 | NSP10/NSP16 | PPI | 20.52% | 24.74% | 46.01% | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID; PHOSPHATE ION; (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid; PHOSPHATE ION; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | XDS | 100.00% | 8.07 | BESSY (14.1) | PHENIX | 2022-02-03 | 36040262 | ZN | - | - | - | N/A | - | - | 2'-O-methyltransferase nsp16; Non-structural protein 10 | 61.4 | - | No | - | 37.8 | 65.8 | 93.8 | - | ||||||
| 7r1u | 2.50 Å | 2022-06-29 | X-ray | MES, GTA, 4IK | SARS-CoV-2 | NSP10/NSP16 | PPI | 23.20% | 25.21% | 60.30% | (2S,5S)-2,6-diamino-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}hexanoic acid; P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID; P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE; (2S,5S)-2,6-diamino-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}hexanoic acid | XDS | 100.00% | 4.64 | ROTATING ANODE () | PHENIX | 2022-02-03 | 36040262 | ZN | - | - | - | N/A | - | - | 2'-O-methyltransferase nsp16; Non-structural protein 10 | 43.1 | - | No | - | 33.2 | 37.7 | 91.0 | - | ||||||
| 7v7m | 2.08 Å | 2022-06-29 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 21.18% | 29.23% | 8.50% | - | CrysalisPro | 99.20% | 15.7 | ROTATING ANODE () | PHENIX | 2021-08-21 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 22.9 | - | No | - | - | 9.2 | 55.3 | 56.4 | - | ||||||
| 7w1s | 2.00 Å | 2022-06-29 | X-ray | - | SARS-CoV-2 | Spike | No functional ligands | 19.87% | 22.39% | 18.20% | - | HKL-3000 | 99.90% | 16.5 | SSRF (BL18U1) | PHENIX | 2021-11-20 | 35663966 | - | - | - | - | N/A | - | - | Spike protein S1; Nanobody Nb-007 | 45.0 | - | No | - | 61.4 | 35.1 | 68.9 | - | |||||||
| 7xh8 | 2.99 Å | 2022-06-29 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-04-07 | 35739114 | - | - | - | - | N/A | - | - | The light chain of ZCB11 antibody; Spike glycoprotein; The heavy chain of ZCB11 antibody | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7xxk | 2.00 Å | 2022-06-29 | X-ray | 5GP, GMP, NA, GUN, K, SCN | SARS-CoV-2 | Nucleocapsid | Functional ligand | 17.83% | 23.19% | 18.40% | POTASSIUM ION; THIOCYANATE ION; GUANOSINE; SODIUM ION; GUANINE; GUANOSINE-5'-MONOPHOSPHATE | GUANOSINE-5'-MONOPHOSPHATE; GUANOSINE; SODIUM ION; GUANINE; POTASSIUM ION; THIOCYANATE ION | XDS | 99.90% | 12.0 | SSRF (BL19U1) | BUSTER | 2022-05-30 | - | NA; K | - | - | - | N/A | - | - | Nucleoprotein - C-terminal domain | 63.7 | - | No | - | - | 53.3 | 77.5 | 71.4 | - | |||||
| 7z59 | 2.00 Å | 2022-06-29 | X-ray | IFO | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 22.23% | 25.16% | 18.80% | (3S)-4-[[2,4-bis(fluoranyl)phenyl]methoxy]-2-methyl-4-oxidanylidene-3-[[(Z)-3-oxidanylidene-2-(2-phenoxyethanoylamino)prop-1-enyl]amino]butane-2-sulfinic acid | (3S)-4-[[2,4-bis(fluoranyl)phenyl]methoxy]-2-methyl-4-oxidanylidene-3-[[(Z)-3-oxidanylidene-2-(2-phenoxyethanoylamino)prop-1-enyl]amino]butane-2-sulfinic acid | xia2 | 100.00% | 7.9 | Diamond (I03) | REFMAC; BUSTER | 2022-03-08 | 35549342 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 27.1 | - | No | - | 33.8 | 52.2 | 43.8 | - | ||||||
| 7zfa | 4.24 Å | 2022-06-29 | X-ray | - | SARS-CoV-2 | Spike | PPI | 23.69% | 27.24% | 33.00% | - | xia2 | 82.90% | 2.6 | Diamond (I03) | PHENIX | 2022-04-01 | 35662412 | - | - | - | - | N/A | - | - | Omi-6 heavy chain; COVOX-150 heavy chain; COVOX-150 light chain; Spike protein S1; Omi-6 light chain | - | - | No | - | 18.2 | - | 46.1 | - | |||||||
| 7zxu | 1.89 Å | 2022-06-29 | X-ray | IPA, PG4, NAG | SARS-CoV-2 | Spike | PPI | 18.07% | 20.90% | 31.30% | ISOPROPYL ALCOHOL; 2-acetamido-2-deoxy-beta-D-glucopyranose | ISOPROPYL ALCOHOL; TETRAETHYLENE GLYCOL; 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 100.00% | 7.7 | Diamond (I03) | PHENIX | 2022-05-23 | 35772405 | - | - | - | - | N/A | - | - | Nanobody C1; Beta-27 light chain; Beta-27 heavy chain; Spike protein S1 | 75.4 | - | No | - | 74.8 | 66.4 | 87.5 | - | ||||||
| 8dce | 2.00 Å | 2022-06-29 | X-ray | - | SARS-CoV-2 | Spike | PPI | 17.72% | 20.30% | N/A | - | XDS | 99.68% | 12.83 | APS (22-ID) | PHENIX; PHENIX | 2022-06-16 | 36103542 | - | - | - | - | N/A | - | - | C144 scFv; Spike protein S1 - RBD SPEEDesign Immunogen 1 (UNP residues 333-526) | 60.7 | - | No | - | 79.6 | 19.1 | 97.2 | - | |||||||
| 5spx | 1.05 Å | 2022-07-06 | X-ray | QIW | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.03% | 15.92% | 4.70% | (5M)-5-(3-ethyl-1H-pyrrolo[2,3-b]pyridin-5-yl)-1,3-dimethyl-1H-pyrazole-4-carboxylic acid | (5M)-5-(3-ethyl-1H-pyrrolo[2,3-b]pyridin-5-yl)-1,3-dimethyl-1H-pyrazole-4-carboxylic acid | XDS | 99.80% | 15.32 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 66.7 | - | No | - | 97.2 | 63.1 | 48.1 | - | ||||||
| 5spy | 1.05 Å | 2022-07-06 | X-ray | QJ0 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.18% | 16.87% | 4.20% | 1-(5-bromo-1H-pyrrolo[2,3-b]pyridin-3-yl)-2-[(1H-tetrazol-5-yl)sulfanyl]ethan-1-one | 1-(5-bromo-1H-pyrrolo[2,3-b]pyridin-3-yl)-2-[(1H-tetrazol-5-yl)sulfanyl]ethan-1-one | XDS | 99.90% | 16.6 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 66.5 | - | No | - | 95.4 | 46.2 | 66.4 | - | ||||||
| 5spz | 1.05 Å | 2022-07-06 | X-ray | QJC | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.07% | 15.95% | 4.90% | (3S)-3-(fluoromethyl)-1-(6-oxo-1,6-dihydropyridazine-4-carbonyl)pyrrolidine-3-carboxylic acid | (3S)-3-(fluoromethyl)-1-(6-oxo-1,6-dihydropyridazine-4-carbonyl)pyrrolidine-3-carboxylic acid | XDS | 99.90% | 14.39 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 72.1 | - | No | - | 97.2 | 59.3 | 64.1 | - | ||||||
| 5sq0 | 1.05 Å | 2022-07-06 | X-ray | QJG | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.80% | 15.61% | 5.30% | (2S,4S)-1-(6-fluoro-2-hydroxyquinoline-4-carbonyl)-4-methylazetidine-2-carboxylic acid | (2S,4S)-1-(6-fluoro-2-hydroxyquinoline-4-carbonyl)-4-methylazetidine-2-carboxylic acid | XDS | 99.80% | 13.3 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 72.7 | - | No | - | 97.6 | 43.4 | 81.0 | - | ||||||
| 5sq1 | 1.05 Å | 2022-07-06 | X-ray | QJO | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.47% | 16.52% | 5.70% | 1-(2-aminopyrimidine-5-sulfonyl)-4,4-difluoro-L-proline | 1-(2-aminopyrimidine-5-sulfonyl)-4,4-difluoro-L-proline | XDS | 99.60% | 12.31 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 65.0 | - | No | - | 96.2 | 36.4 | 72.3 | - | ||||||
| 5sq2 | 1.05 Å | 2022-07-06 | X-ray | QJU | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.95% | 15.84% | 3.80% | 7-fluoro-4-{(3R)-3-[(3-methyl-1,2,4-oxadiazol-5-yl)methyl]piperidin-1-yl}-9H-pyrimido[4,5-b]indole | 7-fluoro-4-{(3R)-3-[(3-methyl-1,2,4-oxadiazol-5-yl)methyl]piperidin-1-yl}-9H-pyrimido[4,5-b]indole | XDS | 99.80% | 19.11 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 64.2 | - | No | - | 97.3 | 34.4 | 71.6 | - | ||||||
| 5sq3 | 1.05 Å | 2022-07-06 | X-ray | QK6 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.46% | 15.18% | 4.60% | [(2R)-4-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)morpholin-2-yl]acetic acid | [(2R)-4-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)morpholin-2-yl]acetic acid | XDS | 99.90% | 16.21 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 63.6 | - | No | - | 98.1 | 49.1 | 54.7 | - | ||||||
| 5sq4 | 1.05 Å | 2022-07-06 | X-ray | QKC | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.15% | 14.73% | 4.10% | 7-fluoro-4-[(3R)-3-(methanesulfonyl)piperidin-1-yl]-9H-pyrimido[4,5-b]indole | 7-fluoro-4-[(3R)-3-(methanesulfonyl)piperidin-1-yl]-9H-pyrimido[4,5-b]indole | XDS | 99.60% | 18.4 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 67.6 | - | No | - | 98.6 | 41.0 | 70.8 | - | ||||||
| 5sq5 | 1.05 Å | 2022-07-06 | X-ray | QKL, QKX | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.59% | 15.53% | 6.10% | (1S,2S)-1-[4-(cyclopropylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid; (1R,2S)-1-[4-(cyclopropylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid | (1R,2S)-1-[4-(cyclopropylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid; (1S,2S)-1-[4-(cyclopropylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.80% | 11.99 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 78.2 | - | No | - | 97.7 | 59.3 | 79.9 | - | ||||||
| 5sq6 | 1.05 Å | 2022-07-06 | X-ray | QL6 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.07% | 17.03% | 5.60% | 3-{[4-(cyclopropylcarbamamido)benzamido]methyl}-1H-indole-2-carboxylic acid | 3-{[4-(cyclopropylcarbamamido)benzamido]methyl}-1H-indole-2-carboxylic acid | XDS | 99.90% | 12.72 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 75.2 | - | No | - | 95.0 | 59.3 | 74.1 | - | ||||||
| 5sq7 | 1.05 Å | 2022-07-06 | X-ray | QM6 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.54% | 19.94% | 5.20% | 3-{[4-(cyclopropylcarbamamido)benzamido]methyl}-1-benzofuran-2-carboxylic acid | 3-{[4-(cyclopropylcarbamamido)benzamido]methyl}-1-benzofuran-2-carboxylic acid | XDS | 99.90% | 13.13 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 42.6 | - | No | - | 82.1 | 26.7 | 52.3 | - | ||||||
| 5sq8 | 1.05 Å | 2022-07-06 | X-ray | QMF | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.24% | 18.44% | 6.90% | 3-{[4-(cyclopropylcarbamamido)benzamido]methyl}-1-benzothiophene-2-carboxylic acid | 3-{[4-(cyclopropylcarbamamido)benzamido]methyl}-1-benzothiophene-2-carboxylic acid | XDS | 99.70% | 9.71 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 51.3 | - | No | - | 90.5 | 26.7 | 60.5 | - | ||||||
| 5sq9 | 1.05 Å | 2022-07-06 | X-ray | QN0, QMO | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.39% | 16.11% | 5.30% | (1S,2S)-4-hydroxy-1-{4-[(propan-2-yl)carbamamido]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid; (1R,2R)-4-hydroxy-1-{4-[(propan-2-yl)carbamamido]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid | (1S,2S)-4-hydroxy-1-{4-[(propan-2-yl)carbamamido]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid; (1R,2R)-4-hydroxy-1-{4-[(propan-2-yl)carbamamido]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.90% | 13.65 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 71.4 | - | No | - | 96.9 | 55.7 | 66.4 | - | ||||||
| 5sqa | 1.05 Å | 2022-07-06 | X-ray | QNF, QNV | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.03% | 15.80% | 3.40% | (1S,2S)-4-hydroxy-1-[(3-oxo-3,4-dihydro-2H-1,4-benzoxazine-7-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid; (1R,2R)-4-hydroxy-1-[(3-oxo-3,4-dihydro-2H-1,4-benzoxazine-7-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid | (1R,2R)-4-hydroxy-1-[(3-oxo-3,4-dihydro-2H-1,4-benzoxazine-7-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid; (1S,2S)-4-hydroxy-1-[(3-oxo-3,4-dihydro-2H-1,4-benzoxazine-7-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.60% | 22.37 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 80.2 | - | No | - | 97.4 | 67.2 | 79.0 | - | ||||||
| 5sqb | 1.05 Å | 2022-07-06 | X-ray | QO3, QOF | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 12.24% | 13.84% | 3.80% | (1S,2S)-1-({6-[(cyclopropylmethyl)amino]pyridine-3-carbonyl}amino)-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid; (1R,2R)-1-({6-[(cyclopropylmethyl)amino]pyridine-3-carbonyl}amino)-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid | (1R,2R)-1-({6-[(cyclopropylmethyl)amino]pyridine-3-carbonyl}amino)-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid; (1S,2S)-1-({6-[(cyclopropylmethyl)amino]pyridine-3-carbonyl}amino)-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.90% | 22.61 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 76.1 | - | No | - | 99.2 | 52.3 | 79.2 | - | ||||||
| 5sqc | 1.05 Å | 2022-07-06 | X-ray | QOR, QP9 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.19% | 15.22% | 4.70% | (1S,2S)-4-hydroxy-1-({5-[(oxan-4-yl)amino]pyrazine-2-carbonyl}amino)-2,3-dihydro-1H-indene-2-carboxylic acid; (1R,2R)-4-hydroxy-1-({5-[(oxan-4-yl)amino]pyrazine-2-carbonyl}amino)-2,3-dihydro-1H-indene-2-carboxylic acid | (1R,2R)-4-hydroxy-1-({5-[(oxan-4-yl)amino]pyrazine-2-carbonyl}amino)-2,3-dihydro-1H-indene-2-carboxylic acid; (1S,2S)-4-hydroxy-1-({5-[(oxan-4-yl)amino]pyrazine-2-carbonyl}amino)-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.80% | 16.54 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 79.6 | - | No | - | 98.1 | 63.1 | 80.2 | - | ||||||
| 5sqd | 1.05 Å | 2022-07-06 | X-ray | QPL, QPX | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.31% | 14.89% | 4.20% | (1S,2S)-4-hydroxy-1-{[2-(hydroxymethyl)-1H-benzimidazole-5-carbonyl]amino}-2,3-dihydro-1H-indene-2-carboxylic acid; (1R,2R)-4-hydroxy-1-{[2-(hydroxymethyl)-1H-benzimidazole-5-carbonyl]amino}-2,3-dihydro-1H-indene-2-carboxylic acid | (1R,2R)-4-hydroxy-1-{[2-(hydroxymethyl)-1H-benzimidazole-5-carbonyl]amino}-2,3-dihydro-1H-indene-2-carboxylic acid; (1S,2S)-4-hydroxy-1-{[2-(hydroxymethyl)-1H-benzimidazole-5-carbonyl]amino}-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.80% | 18.47 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 63.6 | - | No | - | 98.4 | 24.4 | 79.2 | - | ||||||
| 5sqe | 1.05 Å | 2022-07-06 | X-ray | QR0 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.41% | 15.33% | 5.20% | (1S,2S)-4-hydroxy-1-{4-[(1H-imidazol-1-yl)amino]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid | (1S,2S)-4-hydroxy-1-{4-[(1H-imidazol-1-yl)amino]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.90% | 15.7 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 58.1 | - | No | - | 97.9 | 26.7 | 66.1 | - | ||||||
| 5sqf | 1.05 Å | 2022-07-06 | X-ray | QQ9 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.52% | 15.29% | 4.00% | (3R)-1-(1H-pyrrolo[2,3-b]pyridine-4-carbonyl)piperidine-3-carboxylic acid | (3R)-1-(1H-pyrrolo[2,3-b]pyridine-4-carbonyl)piperidine-3-carboxylic acid | XDS | 100.00% | 18.66 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 77.5 | - | No | - | 98.0 | 63.1 | 73.6 | - | ||||||
| 5sqg | 1.05 Å | 2022-07-06 | X-ray | QQI, QQR | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.84% | 15.88% | 6.20% | (1S,2S)-1-{[6-(cyclopropylcarbamamido)pyridine-3-carbonyl]amino}-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid; (1R,2R)-1-{[6-(cyclopropylcarbamamido)pyridine-3-carbonyl]amino}-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid | (1R,2R)-1-{[6-(cyclopropylcarbamamido)pyridine-3-carbonyl]amino}-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid; (1S,2S)-1-{[6-(cyclopropylcarbamamido)pyridine-3-carbonyl]amino}-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.90% | 11.86 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 77.0 | - | No | - | 97.3 | 67.2 | 69.0 | - | ||||||
| 5sqh | 1.05 Å | 2022-07-06 | X-ray | QR6 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.33% | 15.06% | 5.10% | (1S,2S)-1-[2-chloro-4-(cyclopropylcarbamamido)benzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid | (1S,2S)-1-[2-chloro-4-(cyclopropylcarbamamido)benzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.90% | 15.41 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 79.0 | - | No | - | 98.2 | 63.1 | 78.0 | - | ||||||
| 5sqi | 1.05 Å | 2022-07-06 | X-ray | QRI, QRC | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.93% | 15.82% | 7.40% | (1S,2S)-4-hydroxy-1-[4-(methylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid; (1R,2R)-4-hydroxy-1-[4-(methylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid | (1R,2R)-4-hydroxy-1-[4-(methylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid; (1S,2S)-4-hydroxy-1-[4-(methylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.90% | 9.85 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 77.9 | - | No | - | 97.3 | 67.2 | 71.4 | - | ||||||
| 5sqj | 1.05 Å | 2022-07-06 | X-ray | QRU | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.29% | 15.31% | 4.10% | (1R,2R)-1-[4-(cyclopropylcarbamamido)benzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid | (1R,2R)-1-[4-(cyclopropylcarbamamido)benzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.70% | 18.99 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 74.9 | - | No | - | 98.0 | 52.3 | 77.4 | - | ||||||
| 5sqk | 1.05 Å | 2022-07-06 | X-ray | QS6 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.27% | 14.94% | 4.40% | (1R,3S)-3-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]cyclopentan-1-ol | (1R,3S)-3-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]cyclopentan-1-ol | XDS | 99.90% | 17.81 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 72.2 | - | No | - | 98.4 | 59.3 | 63.2 | - | ||||||
| 5sql | 1.05 Å | 2022-07-06 | X-ray | QSL | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.33% | 14.95% | 5.10% | 3-{[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]methyl}-1lambda~6~-thietane-1,1-dione | 3-{[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]methyl}-1lambda~6~-thietane-1,1-dione | XDS | 99.80% | 15.14 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 57.5 | - | No | - | 98.4 | 25.4 | 65.6 | - | ||||||
| 5sqm | 1.05 Å | 2022-07-06 | X-ray | QT0 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.84% | 15.65% | 3.80% | 5-[(3R)-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)piperidin-3-yl]-1,3,4-oxadiazol-2(3H)-one | 5-[(3R)-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)piperidin-3-yl]-1,3,4-oxadiazol-2(3H)-one | XDS | 99.80% | 19.72 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 71.2 | - | No | - | 97.6 | 49.1 | 71.8 | - | ||||||
| 5sqn | 1.05 Å | 2022-07-06 | X-ray | QT6 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.39% | 16.11% | 6.40% | 3-[(3S)-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)piperidin-3-yl]propanoic acid | 3-[(3S)-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)piperidin-3-yl]propanoic acid | XDS | 99.80% | 10.86 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 56.5 | - | No | - | 96.9 | 32.6 | 58.0 | - | ||||||
| 5sqo | 1.05 Å | 2022-07-06 | X-ray | QTF | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.36% | 16.21% | 5.20% | (8R)-8-fluoro-6-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-6-azaspiro[3.4]octane-8-carboxylic acid | (8R)-8-fluoro-6-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-6-azaspiro[3.4]octane-8-carboxylic acid | XDS | 99.80% | 14.06 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 59.9 | - | No | - | 96.7 | 21.5 | 75.9 | - | ||||||
| 5sqp | 1.05 Å | 2022-07-06 | X-ray | QTO | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.30% | 15.90% | 5.70% | (8S)-6-(6-anilinopyrimidin-4-yl)-8-fluoro-6-azaspiro[3.4]octane-8-carboxylic acid | (8S)-6-(6-anilinopyrimidin-4-yl)-8-fluoro-6-azaspiro[3.4]octane-8-carboxylic acid | XDS | 99.80% | 12.45 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 60.9 | - | No | - | 97.2 | 25.4 | 73.6 | - | ||||||
| 5sqq | 1.05 Å | 2022-07-06 | X-ray | QU3 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.11% | 14.61% | 5.40% | 4-(2-amino-7,8-dihydropyrido[4,3-d]pyrimidine-6(5H)-carbonyl)-N-methylfuran-2-sulfonamide | 4-(2-amino-7,8-dihydropyrido[4,3-d]pyrimidine-6(5H)-carbonyl)-N-methylfuran-2-sulfonamide | XDS | 99.80% | 14.81 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 77.1 | - | No | - | 98.7 | 59.3 | 75.8 | - | ||||||
| 5sqr | 1.05 Å | 2022-07-06 | X-ray | QUC | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.26% | 14.90% | 3.80% | (2R,3S)-1-(5-chloro-1H-pyrrolo[2,3-b]pyridine-3-sulfonyl)-2-methylpiperidine-3-carboxylic acid | (2R,3S)-1-(5-chloro-1H-pyrrolo[2,3-b]pyridine-3-sulfonyl)-2-methylpiperidine-3-carboxylic acid | XDS | 99.90% | 20.0 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 62.5 | - | No | - | 98.4 | 46.2 | 55.1 | - | ||||||
| 5sqs | 1.05 Å | 2022-07-06 | X-ray | QVC | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.42% | 15.00% | 5.20% | (4P)-4-[(4M)-4-(3-methyl-1,2,4-oxadiazol-5-yl)pyridin-2-yl]-1H-pyrrolo[2,3-b]pyridine | (4P)-4-[(4M)-4-(3-methyl-1,2,4-oxadiazol-5-yl)pyridin-2-yl]-1H-pyrrolo[2,3-b]pyridine | XDS | 99.80% | 14.44 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 63.3 | - | No | - | 98.3 | 38.5 | 64.5 | - | ||||||
| 5sqt | 1.05 Å | 2022-07-06 | X-ray | QV1, QUR | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.87% | 15.68% | 6.60% | (3R,4R)-4-methyl-1-(2-oxo-2,3-dihydro-1,3-benzoxazole-7-carbonyl)pyrrolidine-3-carboxylic acid; (3S,4S)-4-methyl-1-(2-oxo-2,3-dihydro-1,3-benzoxazole-7-carbonyl)pyrrolidine-3-carboxylic acid | (3S,4S)-4-methyl-1-(2-oxo-2,3-dihydro-1,3-benzoxazole-7-carbonyl)pyrrolidine-3-carboxylic acid; (3R,4R)-4-methyl-1-(2-oxo-2,3-dihydro-1,3-benzoxazole-7-carbonyl)pyrrolidine-3-carboxylic acid | XDS | 99.80% | 10.97 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 75.8 | - | No | - | 97.5 | 59.3 | 73.2 | - | ||||||
| 5squ | 1.05 Å | 2022-07-06 | X-ray | QVL | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.95% | 15.62% | 4.20% | N,3-dimethyl-N-(1H-tetrazol-5-yl)-1H-pyrrolo[2,3-b]pyridine-5-carboxamide | N,3-dimethyl-N-(1H-tetrazol-5-yl)-1H-pyrrolo[2,3-b]pyridine-5-carboxamide | XDS | 99.90% | 17.35 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 67.7 | - | No | - | 97.6 | 49.1 | 64.0 | - | ||||||
| 5sqv | 1.05 Å | 2022-07-06 | X-ray | QVX | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.09% | 16.05% | 3.60% | (1S,2S)-4-hydroxy-1-{4-[(pyridin-3-yl)amino]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid | (1S,2S)-4-hydroxy-1-{4-[(pyridin-3-yl)amino]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.80% | 19.97 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 77.6 | - | No | - | 97.0 | 67.2 | 71.0 | - | ||||||
| 5sqw | 1.05 Å | 2022-07-06 | X-ray | QWC, QW3 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.57% | 15.62% | 3.00% | (1R,2R)-1-[4-(cyclopropylcarbamamido)-2-hydroxybenzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid; (1S,2S)-1-[4-(cyclopropylcarbamamido)-2-hydroxybenzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid | (1S,2S)-1-[4-(cyclopropylcarbamamido)-2-hydroxybenzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid; (1R,2R)-1-[4-(cyclopropylcarbamamido)-2-hydroxybenzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.80% | 27.81 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 77.2 | - | No | - | 97.6 | 55.7 | 80.7 | - | ||||||
| 5sqx | 1.05 Å | 2022-07-06 | X-ray | QWX, QX5 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.07% | 17.43% | 8.50% | (1R,2R)-1-[2-amino-4-(cyclopropylcarbamamido)benzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid; (1S,2S)-1-[2-amino-4-(cyclopropylcarbamamido)benzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid | (1R,2R)-1-[2-amino-4-(cyclopropylcarbamamido)benzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid; (1S,2S)-1-[2-amino-4-(cyclopropylcarbamamido)benzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.90% | 8.1 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 81.1 | - | No | - | 94.0 | 67.2 | 85.4 | - | ||||||
| 5sqy | 1.05 Å | 2022-07-06 | X-ray | QWO | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.99% | 15.83% | 3.10% | (1S,2S)-1-{[4-(cyclopropylcarbamamido)-1,3-benzothiazole-7-carbonyl]amino}-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid | (1S,2S)-1-{[4-(cyclopropylcarbamamido)-1,3-benzothiazole-7-carbonyl]amino}-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.60% | 24.12 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 76.3 | - | No | - | 97.3 | 63.1 | 71.1 | - | ||||||
| 5sqz | 1.05 Å | 2022-07-06 | X-ray | QXC | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.49% | 15.39% | 5.80% | N-[(pyridin-2-yl)methyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine | N-[(pyridin-2-yl)methyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine | XDS | 99.80% | 12.7 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 73.2 | - | No | - | 97.9 | 63.1 | 62.3 | - | ||||||
| 5sr0 | 1.05 Å | 2022-07-06 | X-ray | QY0, QXS | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.06% | 16.03% | 8.40% | (1R,2S)-2-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]cyclohexan-1-ol; (1S,2R)-2-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]cyclohexan-1-ol | (1R,2S)-2-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]cyclohexan-1-ol; (1S,2R)-2-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]cyclohexan-1-ol | XDS | 99.80% | 8.35 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 63.5 | - | No | - | 97.0 | 32.6 | 72.0 | - | ||||||
| 5sr1 | 1.05 Å | 2022-07-06 | X-ray | QXL | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.00% | 14.93% | 4.40% | (3R)-3-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]-1lambda~6~-thiane-1,1-dione | (3R)-3-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]-1lambda~6~-thiane-1,1-dione | XDS | 99.80% | 17.37 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 63.4 | - | No | - | 98.4 | 49.1 | 54.1 | - | ||||||
| 5sr2 | 1.05 Å | 2022-07-06 | X-ray | QYC | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.65% | 15.55% | 3.60% | (1R,2S)-1-[4-(cyclopropylcarbamamido)benzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid | (1R,2S)-1-[4-(cyclopropylcarbamamido)benzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.90% | 20.53 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 73.4 | - | No | - | 97.7 | 52.3 | 73.8 | - | ||||||
| 5sr3 | 1.05 Å | 2022-07-06 | X-ray | QYJ | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.66% | 15.43% | 4.80% | (1S,2S)-1-[4-(cyclopropylcarbamamido)benzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid | (1S,2S)-1-[4-(cyclopropylcarbamamido)benzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.70% | 16.42 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 74.8 | - | No | - | 97.8 | 43.4 | 86.1 | - | ||||||
| 5sr4 | 1.05 Å | 2022-07-06 | X-ray | QYU, QYO | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.00% | 15.57% | 5.30% | (1R,3S)-3-[(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)amino]cyclopentan-1-ol; (1S,3R)-3-[(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)amino]cyclopentan-1-ol | (1R,3S)-3-[(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)amino]cyclopentan-1-ol; (1S,3R)-3-[(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)amino]cyclopentan-1-ol | XDS | 99.70% | 13.77 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 80.1 | - | No | - | 97.7 | 63.1 | 82.3 | - | ||||||
| 5sr5 | 1.05 Å | 2022-07-06 | X-ray | QZ6 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.05% | 14.71% | 4.50% | (2R)-2-{[(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)amino]methyl}-1lambda~6~-thietane-1,1-dione | (2R)-2-{[(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)amino]methyl}-1lambda~6~-thietane-1,1-dione | XDS | 99.70% | 17.15 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 73.0 | - | No | - | 98.6 | 46.2 | 78.0 | - | ||||||
| 5sr6 | 1.05 Å | 2022-07-06 | X-ray | QZF | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.56% | 15.33% | 5.00% | (8R)-8-fluoro-6-(9H-pyrimido[4,5-b]indol-4-yl)-6-azaspiro[3.4]octane-8-carboxylic acid | (8R)-8-fluoro-6-(9H-pyrimido[4,5-b]indol-4-yl)-6-azaspiro[3.4]octane-8-carboxylic acid | XDS | 99.80% | 14.29 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 71.2 | - | No | - | 97.9 | 49.1 | 71.3 | - | ||||||
| 5sr7 | 1.05 Å | 2022-07-06 | X-ray | QZO, QZX | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.37% | 17.44% | 7.90% | (1S,6R,7S)-3-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-3-azabicyclo[4.1.0]heptane-7-carboxylic acid; (1R,6S,7R)-3-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-3-azabicyclo[4.1.0]heptane-7-carboxylic acid | (1S,6R,7S)-3-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-3-azabicyclo[4.1.0]heptane-7-carboxylic acid; (1R,6S,7R)-3-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-3-azabicyclo[4.1.0]heptane-7-carboxylic acid | XDS | 99.60% | 8.51 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 56.4 | - | No | - | 94.0 | 27.9 | 65.4 | - | ||||||
| 5sr8 | 1.10 Å | 2022-07-06 | X-ray | R0A | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.01% | 17.10% | 7.30% | [(6S)-8-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-5-oxa-8-azaspiro[3.5]nonan-6-yl]acetic acid | [(6S)-8-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-5-oxa-8-azaspiro[3.5]nonan-6-yl]acetic acid | XDS | 99.60% | 9.88 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 77.5 | - | No | - | 94.8 | 55.7 | 84.3 | - | ||||||
| 5sr9 | 1.05 Å | 2022-07-06 | X-ray | R0H | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.50% | 15.35% | 4.90% | methyl (3R)-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-3-(hydroxymethyl)pyrrolidine-3-carboxylate | methyl (3R)-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-3-(hydroxymethyl)pyrrolidine-3-carboxylate | XDS | 99.80% | 14.79 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 63.7 | - | No | - | 97.9 | 46.2 | 57.9 | - | ||||||
| 5sra | 1.05 Å | 2022-07-06 | X-ray | R0L | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.76% | 15.70% | 3.80% | (2R)-4-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)morpholine-2-carboxylic acid | (2R)-4-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)morpholine-2-carboxylic acid | XDS | 99.90% | 19.18 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 74.2 | - | No | - | 97.5 | 55.7 | 72.6 | - | ||||||
| 5srb | 1.05 Å | 2022-07-06 | X-ray | R0R, R0W | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.94% | 15.80% | 5.90% | (8S)-8-fluoro-6-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-2-oxa-6-azaspiro[3.4]octane-8-carboxylic acid; (8R)-8-fluoro-6-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-2-oxa-6-azaspiro[3.4]octane-8-carboxylic acid | (8R)-8-fluoro-6-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-2-oxa-6-azaspiro[3.4]octane-8-carboxylic acid; (8S)-8-fluoro-6-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-2-oxa-6-azaspiro[3.4]octane-8-carboxylic acid | XDS | 99.80% | 11.67 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 68.3 | - | No | - | 97.4 | 38.5 | 76.1 | - | ||||||
| 5src | 1.05 Å | 2022-07-06 | X-ray | QIO, QIR | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.56% | 17.54% | 4.50% | (2R)-[(3R)-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)piperidin-3-yl](hydroxy)acetic acid; (2S)-[(3R)-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)piperidin-3-yl](hydroxy)acetic acid | (2R)-[(3R)-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)piperidin-3-yl](hydroxy)acetic acid; (2S)-[(3R)-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)piperidin-3-yl](hydroxy)acetic acid | XDS | 99.80% | 15.42 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 55.4 | - | No | - | 93.7 | 34.4 | 57.4 | - | ||||||
| 5srd | 1.05 Å | 2022-07-06 | X-ray | R8K | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.69% | 16.52% | 5.80% | (8S)-8-fluoro-6-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-N-(methanesulfonyl)-6-azaspiro[3.4]octane-8-carboxamide | (8S)-8-fluoro-6-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-N-(methanesulfonyl)-6-azaspiro[3.4]octane-8-carboxamide | XDS | 99.80% | 12.64 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 80.3 | - | No | - | 96.2 | 63.1 | 84.3 | - | ||||||
| 5sre | 1.05 Å | 2022-07-06 | X-ray | R8R | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.37% | 15.19% | 4.30% | (5R)-7-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-7-azaspiro[3.5]nonane-5-carboxylic acid | (5R)-7-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-7-azaspiro[3.5]nonane-5-carboxylic acid | XDS | 99.70% | 17.12 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 66.3 | - | No | - | 98.1 | 63.1 | 46.4 | - | ||||||
| 5srf | 1.05 Å | 2022-07-06 | X-ray | R8Z | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.85% | 15.60% | 6.10% | (3R)-3-[(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)(methyl)amino]-1lambda~6~-thiane-1,1-dione | (3R)-3-[(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)(methyl)amino]-1lambda~6~-thiane-1,1-dione | XDS | 99.80% | 12.04 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 67.2 | - | No | - | 97.6 | 34.4 | 77.6 | - | ||||||
| 5srg | 1.05 Å | 2022-07-06 | X-ray | R98 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.48% | 15.07% | 5.10% | 7-fluoro-N-methyl-N-[(pyridin-2-yl)methyl]-9H-pyrimido[4,5-b]indol-4-amine | 7-fluoro-N-methyl-N-[(pyridin-2-yl)methyl]-9H-pyrimido[4,5-b]indol-4-amine | XDS | 99.80% | 14.7 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 53.7 | - | No | - | 98.2 | 22.4 | 61.5 | - | ||||||
| 5srh | 1.05 Å | 2022-07-06 | X-ray | TFA, R9F | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.38% | 16.11% | 5.90% | 7-fluoro-9H-pyrimido[4,5-b]indol-4-amine; trifluoroacetic acid | trifluoroacetic acid; 7-fluoro-9H-pyrimido[4,5-b]indol-4-amine | XDS | 99.80% | 12.11 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 60.8 | - | No | - | 96.9 | 22.4 | 76.8 | - | ||||||
| 5sri | 1.05 Å | 2022-07-06 | X-ray | R9F | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.05% | 15.75% | 5.40% | 7-fluoro-9H-pyrimido[4,5-b]indol-4-amine | 7-fluoro-9H-pyrimido[4,5-b]indol-4-amine | XDS | 99.60% | 13.73 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 60.8 | - | No | - | 97.4 | 19.4 | 79.3 | - | ||||||
| 5srj | 1.05 Å | 2022-07-06 | X-ray | R9L | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.38% | 14.90% | 4.40% | 3-{[(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)amino]methyl}-1lambda~6~-thietane-1,1-dione | 3-{[(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)amino]methyl}-1lambda~6~-thietane-1,1-dione | XDS | 99.60% | 17.43 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 72.2 | - | No | - | 98.4 | 55.7 | 66.8 | - | ||||||
| 5srk | 1.05 Å | 2022-07-06 | X-ray | R9U | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 12.93% | 14.56% | 3.70% | (2R)-4-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-N-(methanesulfonyl)morpholine-2-carboxamide | (2R)-4-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-N-(methanesulfonyl)morpholine-2-carboxamide | XDS | 99.80% | 20.7 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 73.7 | - | No | - | 98.7 | 67.2 | 58.6 | - | ||||||
| 5srl | 1.05 Å | 2022-07-06 | X-ray | RA3 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.12% | 14.80% | 4.50% | [(2R,6R)-4-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-6-(hydroxymethyl)morpholin-2-yl]acetic acid | [(2R,6R)-4-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-6-(hydroxymethyl)morpholin-2-yl]acetic acid | XDS | 99.80% | 17.38 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 74.2 | - | No | - | 98.5 | 52.3 | 74.9 | - | ||||||
| 5srm | 1.05 Å | 2022-07-06 | X-ray | RBB | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.32% | 15.06% | 4.50% | [(2R)-6,6-dimethyl-4-(9H-pyrimido[4,5-b]indol-4-yl)morpholin-2-yl]acetic acid | [(2R)-6,6-dimethyl-4-(9H-pyrimido[4,5-b]indol-4-yl)morpholin-2-yl]acetic acid | XDS | 99.80% | 16.54 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 66.8 | - | No | - | 98.2 | 59.3 | 51.2 | - | ||||||
| 5srn | 1.05 Å | 2022-07-06 | X-ray | RBO, RC3 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.22% | 14.95% | 3.50% | [(2S)-4-(9H-pyrimido[4,5-b]indol-4-yl)morpholin-2-yl]acetic acid; [(2R)-4-(9H-pyrimido[4,5-b]indol-4-yl)morpholin-2-yl]acetic acid | [(2R)-4-(9H-pyrimido[4,5-b]indol-4-yl)morpholin-2-yl]acetic acid; [(2S)-4-(9H-pyrimido[4,5-b]indol-4-yl)morpholin-2-yl]acetic acid | XDS | 99.70% | 22.31 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 76.2 | - | No | - | 98.4 | 55.7 | 77.1 | - | ||||||
| 5sro | 1.05 Å | 2022-07-06 | X-ray | RCR, RD6 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.55% | 15.33% | 3.90% | (8S)-6-(7-bromo-9H-pyrimido[4,5-b]indol-4-yl)-8-fluoro-6-azaspiro[3.4]octane-8-carboxylic acid; (8R)-6-(7-bromo-9H-pyrimido[4,5-b]indol-4-yl)-8-fluoro-6-azaspiro[3.4]octane-8-carboxylic acid | (8R)-6-(7-bromo-9H-pyrimido[4,5-b]indol-4-yl)-8-fluoro-6-azaspiro[3.4]octane-8-carboxylic acid; (8S)-6-(7-bromo-9H-pyrimido[4,5-b]indol-4-yl)-8-fluoro-6-azaspiro[3.4]octane-8-carboxylic acid | XDS | 99.70% | 18.97 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 59.8 | - | No | - | 97.9 | 49.1 | 47.0 | - | ||||||
| 5srp | 1.05 Å | 2022-07-06 | X-ray | TFA, RDN | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.17% | 15.04% | 3.90% | trifluoroacetic acid; (8R)-6-(9H-pyrimido[4,5-b]indol-4-yl)-6-azaspiro[3.4]octane-8-carboxylic acid | trifluoroacetic acid; (8R)-6-(9H-pyrimido[4,5-b]indol-4-yl)-6-azaspiro[3.4]octane-8-carboxylic acid | XDS | 99.70% | 19.97 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 71.9 | - | No | - | 98.3 | 59.3 | 62.5 | - | ||||||
| 5srq | 1.05 Å | 2022-07-06 | X-ray | RDU | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.05% | 14.74% | 4.90% | [(6R)-8-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-5-oxa-8-azaspiro[3.5]nonan-6-yl]methanol | [(6R)-8-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-5-oxa-8-azaspiro[3.5]nonan-6-yl]methanol | XDS | 99.80% | 15.4 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 67.6 | - | No | - | 98.5 | 52.3 | 59.5 | - | ||||||
| 5srr | 1.05 Å | 2022-07-06 | X-ray | RF0 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.23% | 14.87% | 3.50% | [(2S,6S)-4-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-6-methoxymorpholin-2-yl]methanol | [(2S,6S)-4-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-6-methoxymorpholin-2-yl]methanol | XDS | 99.70% | 22.37 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 68.9 | - | No | - | 98.4 | 38.5 | 76.3 | - | ||||||
| 5srs | 1.05 Å | 2022-07-06 | X-ray | RFU, RFI | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 12.92% | 14.35% | 4.50% | 3-[(3R)-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)pyrrolidin-3-yl]-1,3-oxazolidin-2-one; 3-[(3S)-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)pyrrolidin-3-yl]-1,3-oxazolidin-2-one | 3-[(3R)-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)pyrrolidin-3-yl]-1,3-oxazolidin-2-one; 3-[(3S)-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)pyrrolidin-3-yl]-1,3-oxazolidin-2-one | XDS | 99.90% | 17.52 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 74.9 | - | No | - | 98.9 | 52.3 | 76.4 | - | ||||||
| 5srt | 1.05 Å | 2022-07-06 | X-ray | RG5 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.07% | 16.68% | 3.80% | 7-fluoro-4-[(2R)-2-(1H-tetrazol-5-yl)morpholin-4-yl]-9H-pyrimido[4,5-b]indole | 7-fluoro-4-[(2R)-2-(1H-tetrazol-5-yl)morpholin-4-yl]-9H-pyrimido[4,5-b]indole | XDS | 99.80% | 19.19 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 64.0 | - | No | - | 95.8 | 36.4 | 70.6 | - | ||||||
| 5sru | 1.05 Å | 2022-07-06 | X-ray | RGF | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.02% | 16.76% | 4.90% | (8S)-6-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-6-azaspiro[3.4]octane-8-carboxylic acid | (8S)-6-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-6-azaspiro[3.4]octane-8-carboxylic acid | XDS | 99.70% | 15.08 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 57.8 | - | No | - | 95.7 | 27.9 | 66.5 | - | ||||||
| 5srv | 1.05 Å | 2022-07-06 | X-ray | RI3 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.35% | 16.52% | 8.20% | (3R,4R)-4-cyclopropyl-3-fluoro-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)pyrrolidine-3-carboxylic acid | (3R,4R)-4-cyclopropyl-3-fluoro-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)pyrrolidine-3-carboxylic acid | XDS | 99.80% | 8.15 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 74.8 | - | No | - | 96.2 | 55.7 | 75.5 | - | ||||||
| 5srw | 1.05 Å | 2022-07-06 | X-ray | RI7 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.02% | 14.58% | 5.90% | methyl [(2S)-4-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)morpholin-2-yl]acetate | methyl [(2S)-4-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)morpholin-2-yl]acetate | XDS | 99.80% | 13.26 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 72.8 | - | No | - | 98.7 | 59.3 | 64.3 | - | ||||||
| 5srx | 1.05 Å | 2022-07-06 | X-ray | RIK | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.48% | 15.13% | 3.70% | 3-[(2R)-4-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)morpholin-2-yl]propanoic acid | 3-[(2R)-4-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)morpholin-2-yl]propanoic acid | XDS | 99.70% | 19.91 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 76.3 | - | No | - | 98.2 | 67.2 | 66.1 | - | ||||||
| 5sry | 1.05 Å | 2022-07-06 | X-ray | RIW | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.35% | 15.05% | 5.80% | 1-[(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)amino]pyrrolidin-2-one | 1-[(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)amino]pyrrolidin-2-one | XDS | 99.90% | 13.17 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 62.9 | - | No | - | 98.2 | 34.4 | 67.9 | - | ||||||
| 5srz | 1.05 Å | 2022-07-06 | X-ray | RIZ, RJ9 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.50% | 16.13% | 6.70% | (1R,2S)-2-[(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)amino]cyclohexane-1-carboxylic acid; (1S,2R)-2-[(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)amino]cyclohexane-1-carboxylic acid | (1R,2S)-2-[(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)amino]cyclohexane-1-carboxylic acid; (1S,2R)-2-[(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)amino]cyclohexane-1-carboxylic acid | XDS | 99.80% | 10.35 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 77.7 | - | No | - | 96.9 | 63.1 | 75.3 | - | ||||||
| 5ss0 | 1.15 Å | 2022-07-06 | X-ray | RJL | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.17% | 15.14% | 5.30% | 3-hydroxy-N-{2-[(5-methoxypyridine-3-carbonyl)amino]ethyl}pyridine-2-carboxamide | 3-hydroxy-N-{2-[(5-methoxypyridine-3-carbonyl)amino]ethyl}pyridine-2-carboxamide | XDS | 99.80% | 15.1 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 76.7 | - | No | - | 98.2 | 55.7 | 78.6 | - | ||||||
| 5ss1 | 1.15 Å | 2022-07-06 | X-ray | RJS | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.14% | 18.71% | 8.60% | (8S)-N-[(4-bromo-3-fluorophenyl)methanesulfonyl]pyrazolo[1,5-a]pyridine-3-carboxamide | (8S)-N-[(4-bromo-3-fluorophenyl)methanesulfonyl]pyrazolo[1,5-a]pyridine-3-carboxamide | XDS | 99.90% | 7.91 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 62.3 | - | No | - | 89.2 | 32.6 | 77.2 | - | ||||||
| 5ss2 | 1.15 Å | 2022-07-06 | X-ray | RK0 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.85% | 15.96% | 5.40% | N-{5-[(3-cyano-4-methylphenyl)sulfamoyl]-4-methyl-1,3-thiazol-2-yl}acetamide | N-{5-[(3-cyano-4-methylphenyl)sulfamoyl]-4-methyl-1,3-thiazol-2-yl}acetamide | XDS | 99.70% | 17.3 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 70.1 | - | No | - | 97.1 | 46.2 | 72.6 | - | ||||||
| 5ss3 | 1.15 Å | 2022-07-06 | X-ray | RK9 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.54% | 17.96% | 10.10% | N-{5-[(3-cyanophenyl)sulfamoyl]-4-methyl-1,3-thiazol-2-yl}propanamide | N-{5-[(3-cyanophenyl)sulfamoyl]-4-methyl-1,3-thiazol-2-yl}propanamide | XDS | 99.90% | 7.65 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 63.3 | - | No | - | 92.3 | 32.6 | 76.5 | - | ||||||
| 5ss4 | 1.15 Å | 2022-07-06 | X-ray | RKI | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.24% | 15.89% | 6.80% | (3-{[(thieno[3,2-d]pyrimidine-4-carbonyl)amino]methyl}phenyl)acetic acid | (3-{[(thieno[3,2-d]pyrimidine-4-carbonyl)amino]methyl}phenyl)acetic acid | XDS | 99.90% | 13.23 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 50.8 | - | No | - | 97.2 | 19.4 | 59.8 | - | ||||||
| 5ss5 | 1.15 Å | 2022-07-06 | X-ray | RKU | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.00% | 16.07% | 4.90% | (3S)-3-(4-bromophenyl)-3-[(6-fluoro-1H-benzimidazole-4-carbonyl)amino]propanoic acid | (3S)-3-(4-bromophenyl)-3-[(6-fluoro-1H-benzimidazole-4-carbonyl)amino]propanoic acid | XDS | 99.60% | 15.66 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 64.0 | - | No | - | 97.0 | 32.6 | 73.2 | - | ||||||
| 5ss6 | 1.15 Å | 2022-07-06 | X-ray | RL5 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.77% | 15.79% | 6.60% | 1-(2-{[2-(ethylamino)-1,3-thiazole-5-carbonyl]amino}ethyl)-1H-imidazole-4-carboxylic acid | 1-(2-{[2-(ethylamino)-1,3-thiazole-5-carbonyl]amino}ethyl)-1H-imidazole-4-carboxylic acid | XDS | 99.80% | 12.32 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 72.2 | - | No | - | 97.4 | 49.1 | 74.3 | - | ||||||
| 5ss7 | 1.15 Å | 2022-07-06 | X-ray | RL9 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 12.78% | 14.60% | 6.30% | 4-fluoro-3-{[(1H-indole-5-carbonyl)amino]methyl}benzoic acid | 4-fluoro-3-{[(1H-indole-5-carbonyl)amino]methyl}benzoic acid | XDS | 99.90% | 14.91 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 76.9 | - | No | - | 98.7 | 59.3 | 75.2 | - | ||||||
| 5ss8 | 1.15 Å | 2022-07-06 | X-ray | RLN | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 12.85% | 14.98% | 4.30% | (4-{[(thieno[3,2-b]pyridine-7-carbonyl)amino]methyl}phenyl)acetic acid | (4-{[(thieno[3,2-b]pyridine-7-carbonyl)amino]methyl}phenyl)acetic acid | XDS | 99.90% | 19.52 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 78.1 | - | No | - | 98.3 | 55.7 | 82.8 | - | ||||||
| 5ss9 | 1.15 Å | 2022-07-06 | X-ray | RLU | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.47% | 16.65% | 5.50% | (3R)-3-(4-bromophenyl)-3-[(7H-pyrrolo[2,3-d]pyrimidine-4-carbonyl)amino]propanoic acid | (3R)-3-(4-bromophenyl)-3-[(7H-pyrrolo[2,3-d]pyrimidine-4-carbonyl)amino]propanoic acid | XDS | 99.60% | 14.01 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 51.4 | - | No | - | 95.9 | 16.8 | 65.0 | - | ||||||
| 5ssa | 1.15 Å | 2022-07-06 | X-ray | RM6 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.78% | 17.17% | 7.40% | [(1r,3r)-3-{[(thieno[2,3-c]pyridine-5-carbonyl)amino]methyl}cyclobutyl]acetic acid | [(1r,3r)-3-{[(thieno[2,3-c]pyridine-5-carbonyl)amino]methyl}cyclobutyl]acetic acid | XDS | 99.90% | 9.93 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 67.6 | - | No | - | 94.7 | 32.6 | 83.0 | - | ||||||
| 5ssb | 1.15 Å | 2022-07-06 | X-ray | RMU | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.57% | 15.67% | 6.10% | 4-[(6-chloro-5-cyanopyridin-3-yl)sulfamoyl]-5-methylfuran-2-carboxamide | 4-[(6-chloro-5-cyanopyridin-3-yl)sulfamoyl]-5-methylfuran-2-carboxamide | XDS | 99.80% | 12.41 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 75.0 | - | No | - | 97.6 | 46.2 | 84.0 | - | ||||||
| 5ssc | 1.15 Å | 2022-07-06 | X-ray | RNC | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.54% | 16.65% | 7.00% | [3-(5-hydroxy-1,2,4-oxadiazol-3-yl)azetidin-1-yl][5-(methylamino)pyrazin-2-yl]methanone | [3-(5-hydroxy-1,2,4-oxadiazol-3-yl)azetidin-1-yl][5-(methylamino)pyrazin-2-yl]methanone | XDS | 99.80% | 10.91 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 63.3 | - | No | - | 95.9 | 34.4 | 71.0 | - | ||||||
| 5ssd | 1.15 Å | 2022-07-06 | X-ray | RNL | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.81% | 15.80% | 7.70% | (1R,2S)-2-({2-[(4S)-7-methyl-8-oxo-7,8-dihydro[1,2,4]triazolo[4,3-a]pyrazin-3-yl]ethyl}carbamoyl)cyclopropane-1-carboxylic acid | (1R,2S)-2-({2-[(4S)-7-methyl-8-oxo-7,8-dihydro[1,2,4]triazolo[4,3-a]pyrazin-3-yl]ethyl}carbamoyl)cyclopropane-1-carboxylic acid | XDS | 99.50% | 10.95 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 80.3 | - | No | - | 97.4 | 59.3 | 87.3 | - | ||||||
| 5sse | 1.15 Å | 2022-07-06 | X-ray | ROO | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 12.96% | 14.89% | 4.80% | 2-methyl-5-{[(9H-pyrimido[4,5-b]indol-4-yl)amino]methyl}furan-3-carboxylic acid | 2-methyl-5-{[(9H-pyrimido[4,5-b]indol-4-yl)amino]methyl}furan-3-carboxylic acid | XDS | 99.70% | 19.54 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 71.9 | - | No | - | 98.4 | 46.2 | 75.5 | - | ||||||
| 5ssf | 1.15 Å | 2022-07-06 | X-ray | RP0 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.08% | 16.52% | 7.30% | 4-[(4-bromo-3-cyanophenyl)sulfamoyl]-5-methylfuran-2-carboxamide | 4-[(4-bromo-3-cyanophenyl)sulfamoyl]-5-methylfuran-2-carboxamide | XDS | 99.60% | 10.41 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 71.0 | - | No | - | 96.2 | 43.4 | 78.3 | - | ||||||
| 5ssg | 1.15 Å | 2022-07-06 | X-ray | RPU | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.91% | 16.26% | 7.50% | (3R)-3-(2H-1,3-benzodioxol-5-yl)-3-[(2R)-3-(furan-2-yl)-2-methylpropanamido]propanoic acid | (3R)-3-(2H-1,3-benzodioxol-5-yl)-3-[(2R)-3-(furan-2-yl)-2-methylpropanamido]propanoic acid | XDS | 99.30% | 10.23 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 57.9 | - | No | - | 96.7 | 34.4 | 59.3 | - | ||||||
| 5ssh | 1.15 Å | 2022-07-06 | X-ray | RQ8 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.19% | 14.93% | 4.70% | 3-(5-bromopyridin-3-yl)-N-[5-(1,1-difluoroethyl)pyridine-3-carbonyl]-L-alanine | 3-(5-bromopyridin-3-yl)-N-[5-(1,1-difluoroethyl)pyridine-3-carbonyl]-L-alanine | XDS | 99.90% | 19.67 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 56.5 | - | No | - | 98.4 | 36.4 | 52.8 | - | ||||||
| 5ssi | 1.15 Å | 2022-07-06 | X-ray | RQC | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.41% | 15.20% | 5.80% | (3R)-1-[3-(1-methyl-4-oxo-4,5-dihydro-1H-pyrazolo[3,4-d]pyrimidin-6-yl)propanoyl]pyrrolidine-3-carboxylic acid | (3R)-1-[3-(1-methyl-4-oxo-4,5-dihydro-1H-pyrazolo[3,4-d]pyrimidin-6-yl)propanoyl]pyrrolidine-3-carboxylic acid | XDS | 99.50% | 14.92 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 73.4 | - | No | - | 98.1 | 52.3 | 73.3 | - | ||||||
| 5ssj | 1.15 Å | 2022-07-06 | X-ray | RQI | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 12.75% | 14.63% | 3.70% | 2-{2-[(6-fluoro-1H-benzimidazole-5-carbonyl)amino]ethyl}-1,3-thiazole-4-carboxylic acid | 2-{2-[(6-fluoro-1H-benzimidazole-5-carbonyl)amino]ethyl}-1,3-thiazole-4-carboxylic acid | XDS | 99.50% | 24.1 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 60.0 | - | No | - | 98.6 | 49.1 | 46.8 | - | ||||||
| 5ssk | 1.15 Å | 2022-07-06 | X-ray | RQR | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 12.49% | 14.46% | 4.80% | (1S,4R)-4-[(thieno[2,3-d]pyrimidine-4-carbonyl)amino]cyclopent-2-ene-1-carboxylic acid | (1S,4R)-4-[(thieno[2,3-d]pyrimidine-4-carbonyl)amino]cyclopent-2-ene-1-carboxylic acid | XDS | 99.60% | 24.47 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 67.5 | - | No | - | 98.8 | 38.5 | 72.8 | - | ||||||
| 5ssl | 1.15 Å | 2022-07-06 | X-ray | RR3 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.12% | 16.31% | 7.10% | 1-[4-(cyanomethyl)phenyl]-N-(1-methyl-1H-pyrazolo[4,3-d]pyrimidin-7-yl)methanesulfonamide | 1-[4-(cyanomethyl)phenyl]-N-(1-methyl-1H-pyrazolo[4,3-d]pyrimidin-7-yl)methanesulfonamide | XDS | 99.70% | 11.7 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 65.9 | - | No | - | 96.6 | 26.7 | 83.6 | - | ||||||
| 5ssm | 1.15 Å | 2022-07-06 | X-ray | RS0, RRF | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.82% | 15.61% | 4.20% | (1R,2R)-1-[(1H-benzimidazole-5-carbonyl)amino]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid; (1S,2S)-1-[(1H-benzimidazole-5-carbonyl)amino]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid | (1S,2S)-1-[(1H-benzimidazole-5-carbonyl)amino]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid; (1R,2R)-1-[(1H-benzimidazole-5-carbonyl)amino]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.40% | 21.54 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 65.0 | - | No | - | 97.6 | 36.4 | 71.0 | - | ||||||
| 5ssn | 1.15 Å | 2022-07-06 | X-ray | RS9 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.40% | 15.21% | 4.50% | (1R,2S)-4-hydroxy-1-{[2-(hydroxymethyl)-1H-benzimidazole-5-carbonyl]amino}-2,3-dihydro-1H-indene-2-carboxylic acid | (1R,2S)-4-hydroxy-1-{[2-(hydroxymethyl)-1H-benzimidazole-5-carbonyl]amino}-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.40% | 19.13 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 58.4 | - | No | - | 98.1 | 24.4 | 68.9 | - | ||||||
| 5sso | 1.15 Å | 2022-07-06 | X-ray | RT5, RSR | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.65% | 15.66% | 5.70% | (8S)-6-(2-amino-7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-8-fluoro-6-azaspiro[3.4]octane-8-carboxylic acid; (8R)-6-(2-amino-7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-8-fluoro-6-azaspiro[3.4]octane-8-carboxylic acid | (8S)-6-(2-amino-7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-8-fluoro-6-azaspiro[3.4]octane-8-carboxylic acid; (8R)-6-(2-amino-7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-8-fluoro-6-azaspiro[3.4]octane-8-carboxylic acid | XDS | 99.70% | 14.95 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 59.6 | - | No | - | 97.6 | 38.5 | 57.6 | - | ||||||
| 5ssp | 1.15 Å | 2022-07-06 | X-ray | RTI, RTU | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.15% | 14.95% | 4.40% | (1R,2R)-4-hydroxy-1-[(1H-indole-5-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid; (1S,2S)-4-hydroxy-1-[(1H-indole-5-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid | (1S,2S)-4-hydroxy-1-[(1H-indole-5-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid; (1R,2R)-4-hydroxy-1-[(1H-indole-5-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.40% | 19.66 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 78.8 | - | No | - | 98.4 | 49.1 | 91.4 | - | ||||||
| 5ssq | 1.15 Å | 2022-07-06 | X-ray | RV3 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.31% | 15.05% | 4.30% | 3-{[(1H-benzimidazole-5-carbonyl)amino]methyl}-1H-indole-2-carboxylic acid | 3-{[(1H-benzimidazole-5-carbonyl)amino]methyl}-1H-indole-2-carboxylic acid | XDS | 99.40% | 20.38 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 66.1 | - | No | - | 98.2 | 29.3 | 79.6 | - | ||||||
| 5ssr | 1.15 Å | 2022-07-06 | X-ray | RVF | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 12.94% | 14.70% | 5.10% | 3-{[(2-hydroxy-1H-benzimidazole-5-carbonyl)amino]methyl}-1H-indole-2-carboxylic acid | 3-{[(2-hydroxy-1H-benzimidazole-5-carbonyl)amino]methyl}-1H-indole-2-carboxylic acid | XDS | 98.80% | 16.78 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 73.7 | - | No | - | 98.6 | 55.7 | 70.4 | - | ||||||
| 7vbe | 1.59 Å | 2022-07-06 | X-ray | - | SARS-CoV-2 | Nucleocapsid | PPI | 20.30% | 22.64% | 7.60% | - | XDS | 99.80% | 8.4 | SSRF (BL17U1) | PHENIX | 2021-08-31 | - | - | - | - | - | N/A | - | - | Nucleoprotein - dimerization domain | 60.3 | - | No | - | - | 58.8 | 70.4 | 65.9 | - | ||||||
| 7vbf | 1.30 Å | 2022-07-06 | X-ray | - | SARS-CoV-2 | Nucleocapsid | PPI | 18.50% | 20.39% | N/A | - | XDS | 99.10% | 19.3 | SSRF (BL17U1) | PHENIX | 2021-08-31 | - | - | - | - | - | N/A | - | - | Nucleoprotein - dimerization domain | 74.6 | - | No | - | - | 79.0 | 59.6 | 88.1 | - | ||||||
| 7x1m | 2.74 Å | 2022-07-06 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-02-24 | 35777362 | - | - | - | - | N/A | - | - | BD-604 Fab heavy chain; S309 Fab light chain; S309 Fab heavy chain; Spike protein S1 - RBD; S304 Fab light chain; S304 Fab haavy chain; BD-604 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7x6j | 1.50 Å | 2022-07-06 | X-ray | QNC | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.97% | 20.36% | 10.00% | quinoline-2-carboxylic acid | quinoline-2-carboxylic acid | HKL-2000 | 98.60% | 7.4 | SSRF (BL18U1) | PHENIX | 2022-03-07 | 35709506 | - | - | - | - | N/A | - | - | 3C-like proteinase | 60.3 | - | No | - | 79.2 | 28.4 | 87.5 | - | ||||||
| 7x6k | 2.34 Å | 2022-07-06 | X-ray | 9FF | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 23.24% | 27.04% | 16.70% | 1H-indole-2-carbaldehyde | 1H-indole-2-carbaldehyde | XDS | 96.90% | 12.0 | SSRF (BL02U1) | PHENIX | 2022-03-07 | 35709506 | - | - | - | - | N/A | - | - | 3C-like proteinase | 58.2 | - | No | - | 19.3 | 78.5 | 93.1 | - | ||||||
| 7xaz | 3.00 Å | 2022-07-06 | X-ray | NAG | SARS-CoV-2 | Spike | PPI | 19.76% | 23.02% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.90% | 11.3 | SSRF (BL02U1) | PHENIX | 2022-03-19 | 35809570 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike protein S1 - Omicron RBD | 58.7 | - | No | - | 54.8 | 92.6 | 44.5 | - | ||||||
| 7xb0 | 2.90 Å | 2022-07-06 | X-ray | NAG | SARS-CoV-2 | Spike | PPI | 21.56% | 25.52% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 96.50% | 13.3 | SSRF (BL10U2) | PHENIX | 2022-03-19 | 35809570 | ZN | - | - | - | N/A | - | - | Spike protein S1 - Omicron BA.2 RBD; Angiotensin-converting enzyme 2 | 9.4 | - | No | - | 30.5 | 20.3 | 35.7 | - | ||||||
| 7xb1 | 2.70 Å | 2022-07-06 | X-ray | NAG | SARS-CoV-2 | Spike | PPI | 21.40% | 24.80% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.80% | 16.6 | SSRF (BL19U1) | PHENIX | 2022-03-19 | 35809570 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike protein S1 - BA.3 RBD | 12.3 | - | No | - | 37.0 | 23.0 | 35.4 | - | ||||||
| 7y42 | 3.45 Å | 2022-07-06 | Cryo-EM | NAG, REA | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; RETINOIC ACID | 2-acetamido-2-deoxy-beta-D-glucopyranose; RETINOIC ACID | - | - | () | 2022-06-13 | 35862773 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8cwu | 1.71 Å | 2022-07-06 | X-ray | - | SARS-CoV-2 | Spike | PPI | 18.51% | 20.32% | 10.30% | - | HKL-2000 | 98.10% | 5.2 | APS (23-ID-D) | PHENIX | 2022-05-19 | 35738279 | - | - | - | - | N/A | - | - | VHH 1-21; Spike protein S1 - Receptor binding domain, UNP residues 333-530 | 48.2 | - | No | - | 79.4 | 25.6 | 66.5 | - | |||||||
| 8cwv | 2.51 Å | 2022-07-06 | X-ray | - | SARS-CoV-2 | Spike | PPI | 23.77% | 26.40% | 14.70% | - | HKL-2000 | 84.00% | 4.3 | APS (23-ID-D) | PHENIX | 2022-05-19 | 35738279 | - | - | - | - | N/A | - | - | CC12.1 Fab light chain; CC12.1 Fab heavy chain; Spike protein S1 - Receptor binding domain, UNP residues 333-530; VHH 2-31 | 24.2 | - | No | - | 23.6 | 54.8 | 45.3 | - | |||||||
| 8cxn | 2.90 Å | 2022-07-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-05-22 | 35738279 | - | - | - | - | N/A | - | - | Spike glycoprotein; pan-sarbecovirus nanobody 2-57 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 8cxq | 2.30 Å | 2022-07-06 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX; phenix.real_space_refine | 2022-05-22 | 35738279 | - | - | - | - | N/A | - | - | pan-sarbecovirus nanobody 1-22; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8cy6 | 3.20 Å | 2022-07-06 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-05-23 | 35738279 | - | - | - | - | N/A | - | - | Spike glycoprotein; pan-sarbecovirus nanobody 2-65 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8cy7 | 2.90 Å | 2022-07-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-05-23 | 35738279 | - | - | - | - | N/A | - | - | pan-sarbecovirus nanobody 2-38; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 8cy9 | 2.90 Å | 2022-07-06 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-05-23 | 35738279 | - | - | - | - | N/A | - | - | Spike glycoprotein; pan-sarbecovirus nanobody 1-23 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8cya | 2.70 Å | 2022-07-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-05-23 | 35738279 | - | - | - | - | N/A | - | - | Spike glycoprotein; pan-sarbecovirus nanobody 2-67 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 8cyb | 2.70 Å | 2022-07-06 | Cryo-EM | NAG, AH2 | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 1-deoxy-alpha-D-mannopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose; 1-deoxy-alpha-D-mannopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-05-23 | 35738279 | - | - | - | - | N/A | - | - | Spike glycoprotein; pan-sarbecovirus nanobody 1-8 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 8cyc | 2.90 Å | 2022-07-06 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-05-23 | 35738279 | - | - | - | - | N/A | - | - | Spike glycoprotein; pan-sarbecovirus nanobody 2-34 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8cyd | 2.60 Å | 2022-07-06 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-05-23 | 35738279 | - | - | - | - | N/A | - | - | Spike glycoprotein; pan-sarbecovirus nanobody 2-45 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8cyj | 3.60 Å | 2022-07-06 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | phenix.real_space_refine; PHENIX | 2022-05-23 | 35738279 | - | - | - | - | N/A | - | - | pan-sarbecovirus nanobody 2-67; pan-sarbecovirus nanobody 2-62; pan-sarbecovirus nanobody 2-10; pan-sarbecovirus nanobody 1-25; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dcc | 2.60 Å | 2022-07-06 | X-ray | NAG | SARS-CoV-2 | Spike | PPI | 22.05% | 27.22% | 18.70% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 100.00% | 8.2 | APS (23-ID-D) | PHENIX | 2022-06-16 | 36103542 | - | - | - | - | N/A | - | - | P2B-2F6 Fab light chain; Spike protein S1 - RBD SPEEDesign Immunogen 3 (UNP residues 333-526); P2B-2F6 Fab heavy chain | 33.8 | - | No | - | 18.3 | 72.3 | 53.0 | - | ||||||
| 8dgy | 1.65 Å | 2022-07-06 | X-ray | SD6, S9U | MERS-CoV | NSP5 (3CLpro) | Functional ligand | 16.37% | 20.23% | 4.80% | [(1~{R},2~{R})-2-(cyclohexylmethyl)cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate; [(1~{R},2~{R})-2-(cyclohexylmethyl)cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | [(1~{R},2~{R})-2-(cyclohexylmethyl)cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate; [(1~{R},2~{R})-2-(cyclohexylmethyl)cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | XDS | 96.90% | 12.9 | NSLS-II (19-ID) | PHENIX | 2022-06-24 | 36654747 | - | - | - | - | N/A | - | - | 3C-like proteinase | 79.8 | - | No | - | 80.1 | 88.6 | 73.4 | - | ||||||
| 5soi | 1.05 Å | 2022-07-13 | X-ray | WYY, WVG | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.79% | 15.57% | 3.10% | 3-[(3S)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]propanoic acid; 3-[(3R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]propanoic acid | 3-[(3S)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]propanoic acid; 3-[(3R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]propanoic acid | XDS | 99.80% | 24.07 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 67.5 | - | No | - | 97.7 | 49.1 | 63.4 | - | ||||||
| 5soj | 1.05 Å | 2022-07-13 | X-ray | RWQ | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.43% | 15.01% | 5.10% | [(3R)-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)piperidin-3-yl]methanol | [(3R)-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)piperidin-3-yl]methanol | XDS | 99.70% | 14.46 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 57.5 | - | No | - | 98.3 | 26.7 | 64.6 | - | ||||||
| 5sok | 1.05 Å | 2022-07-13 | X-ray | RVO | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.02% | 15.91% | 7.60% | 5-chloro-6-{(3R)-3-[(pyridin-4-yl)oxy]pyrrolidin-1-yl}pyrimidin-4-amine | 5-chloro-6-{(3R)-3-[(pyridin-4-yl)oxy]pyrrolidin-1-yl}pyrimidin-4-amine | XDS | 99.80% | 9.68 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 76.6 | - | No | - | 97.2 | 63.1 | 71.9 | - | ||||||
| 5sol | 1.05 Å | 2022-07-13 | X-ray | WVM | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.62% | 16.28% | 3.00% | (8S)-8-fluoro-6-(6-{[(2R)-2-hydroxypropyl]amino}pyrimidin-4-yl)-6-azaspiro[3.4]octane-8-carboxylic acid | (8S)-8-fluoro-6-(6-{[(2R)-2-hydroxypropyl]amino}pyrimidin-4-yl)-6-azaspiro[3.4]octane-8-carboxylic acid | XDS | 99.70% | 23.77 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 70.3 | - | No | - | 96.6 | 46.2 | 73.4 | - | ||||||
| 5som | 1.05 Å | 2022-07-13 | X-ray | WVY | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.75% | 15.56% | 6.60% | [(3S)-2-oxopiperidin-3-yl]methyl [4-(1H-pyrazol-1-yl)phenyl]acetate | [(3S)-2-oxopiperidin-3-yl]methyl [4-(1H-pyrazol-1-yl)phenyl]acetate | XDS | 100.00% | 11.72 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 77.5 | - | No | - | 97.7 | 67.2 | 70.0 | - | ||||||
| 5son | 1.05 Å | 2022-07-13 | X-ray | WW1 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.65% | 17.83% | 5.90% | 3-{[(2R)-2-phenylpropyl]sulfanyl}-7H-[1,2,4]triazolo[4,3-b][1,2,4]triazole | 3-{[(2R)-2-phenylpropyl]sulfanyl}-7H-[1,2,4]triazolo[4,3-b][1,2,4]triazole | XDS | 99.90% | 11.39 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 67.5 | - | No | - | 92.7 | 46.2 | 71.3 | - | ||||||
| 5soo | 1.05 Å | 2022-07-13 | X-ray | WW4 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.08% | 15.74% | 4.00% | 4-{(3R)-3-[(1,3-thiazol-2-yl)methyl]pyrrolidin-1-yl}-7H-pyrrolo[2,3-d]pyrimidine | 4-{(3R)-3-[(1,3-thiazol-2-yl)methyl]pyrrolidin-1-yl}-7H-pyrrolo[2,3-d]pyrimidine | XDS | 99.70% | 18.27 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 56.6 | - | No | - | 97.4 | 21.5 | 68.7 | - | ||||||
| 5sop | 1.05 Å | 2022-07-13 | X-ray | RYI | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.66% | 17.36% | 3.70% | (5R)-7-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-2lambda~6~-thia-7-azaspiro[4.5]decane-2,2-dione | (5R)-7-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-2lambda~6~-thia-7-azaspiro[4.5]decane-2,2-dione | XDS | 100.00% | 19.46 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 46.5 | - | No | - | 94.2 | 16.5 | 57.8 | - | ||||||
| 5soq | 1.05 Å | 2022-07-13 | X-ray | WWJ | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.74% | 15.58% | 4.80% | 5-ethyl-4-{(3R)-3-[(3-methyl-1,2,4-oxadiazol-5-yl)methyl]piperidin-1-yl}-7H-pyrrolo[2,3-d]pyrimidine | 5-ethyl-4-{(3R)-3-[(3-methyl-1,2,4-oxadiazol-5-yl)methyl]piperidin-1-yl}-7H-pyrrolo[2,3-d]pyrimidine | XDS | 99.70% | 15.15 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 56.5 | - | No | - | 97.7 | 16.8 | 73.0 | - | ||||||
| 5sor | 1.05 Å | 2022-07-13 | X-ray | WWP | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.80% | 15.58% | 4.40% | 3-{[3-(trifluoromethyl)phenyl]methyl}-3H-purin-6-amine | 3-{[3-(trifluoromethyl)phenyl]methyl}-3H-purin-6-amine | XDS | 99.90% | 17.5 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 49.3 | - | No | - | 97.7 | 18.8 | 57.3 | - | ||||||
| 5sos | 1.05 Å | 2022-07-13 | X-ray | WWS | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.38% | 15.06% | 3.30% | 3-[(5-chloropyridin-2-yl)methyl]-3H-purin-6-amine | 3-[(5-chloropyridin-2-yl)methyl]-3H-purin-6-amine | XDS | 99.90% | 23.24 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 71.3 | - | No | - | 98.2 | 38.5 | 82.0 | - | ||||||
| 5sot | 1.05 Å | 2022-07-13 | X-ray | S1O, WX4 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.57% | 15.42% | 3.30% | {1-[(3R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]-1H-1,2,3-triazol-4-yl}methanol; {1-[(3S)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]-1H-1,2,3-triazol-4-yl}methanol | {1-[(3S)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]-1H-1,2,3-triazol-4-yl}methanol; {1-[(3R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]-1H-1,2,3-triazol-4-yl}methanol | XDS | 99.80% | 22.57 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 74.1 | - | No | - | 97.8 | 63.1 | 64.5 | - | ||||||
| 5sou | 1.05 Å | 2022-07-13 | X-ray | RZ9, WX7 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.65% | 15.21% | 3.00% | 5-[(3S)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]-1,3,4-oxadiazol-2(3H)-one; 5-[(3R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]-1,3,4-oxadiazol-2(3H)-one | 5-[(3S)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]-1,3,4-oxadiazol-2(3H)-one; 5-[(3R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]-1,3,4-oxadiazol-2(3H)-one | XDS | 99.80% | 24.48 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 71.4 | - | No | - | 98.1 | 55.7 | 65.2 | - | ||||||
| 5sov | 1.05 Å | 2022-07-13 | X-ray | WXD, WXA | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.93% | 15.70% | 2.80% | 5-ethyl-4-[(3S)-3-(methylsulfonyl)piperidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine; 5-ethyl-4-[(3R)-3-(methylsulfonyl)piperidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine | 5-ethyl-4-[(3S)-3-(methylsulfonyl)piperidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine; 5-ethyl-4-[(3R)-3-(methylsulfonyl)piperidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine | XDS | 99.90% | 26.28 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 72.3 | - | No | - | 97.5 | 49.1 | 74.4 | - | ||||||
| 5sow | 1.05 Å | 2022-07-13 | X-ray | WXG | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.62% | 17.70% | 4.00% | 1-{2-[(9H-purin-6-yl)sulfanyl]ethyl}pyrrolidin-2-one | 1-{2-[(9H-purin-6-yl)sulfanyl]ethyl}pyrrolidin-2-one | XDS | 99.80% | 17.76 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 75.5 | - | No | - | 93.2 | 67.2 | 68.8 | - | ||||||
| 5sox | 1.05 Å | 2022-07-13 | X-ray | WXJ | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 12.03% | 13.56% | 3.50% | 4-[2-(6-amino-3H-purin-3-yl)ethoxy]benzonitrile | 4-[2-(6-amino-3H-purin-3-yl)ethoxy]benzonitrile | XDS | 100.00% | 25.01 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 77.7 | - | No | - | 99.4 | 59.3 | 76.8 | - | ||||||
| 5soy | 1.05 Å | 2022-07-13 | X-ray | WXS | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 16.28% | 18.30% | 6.90% | 4-methyl-5-{[(9H-purin-6-yl)sulfanyl]methyl}-2H-1,3-dioxol-2-one | 4-methyl-5-{[(9H-purin-6-yl)sulfanyl]methyl}-2H-1,3-dioxol-2-one | XDS | 99.90% | 9.37 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 62.1 | - | No | - | 91.0 | 43.4 | 64.5 | - | ||||||
| 5soz | 1.05 Å | 2022-07-13 | X-ray | WXY | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.06% | 16.20% | 5.00% | (1-azaspiro[4.5]decan-1-yl)(7H-pyrrolo[2,3-d]pyrimidin-4-yl)methanone | (1-azaspiro[4.5]decan-1-yl)(7H-pyrrolo[2,3-d]pyrimidin-4-yl)methanone | XDS | 99.80% | 14.57 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 55.0 | - | No | - | 96.8 | 23.4 | 64.6 | - | ||||||
| 5sp0 | 1.05 Å | 2022-07-13 | X-ray | WY7 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 12.95% | 14.65% | 3.40% | 3-{[5-(furan-2-yl)-1,2-oxazol-3-yl]methyl}-3H-purin-6-amine | 3-{[5-(furan-2-yl)-1,2-oxazol-3-yl]methyl}-3H-purin-6-amine | XDS | 99.90% | 24.56 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 71.2 | - | No | - | 98.6 | 46.2 | 73.5 | - | ||||||
| 5sp1 | 1.03 Å | 2022-07-13 | X-ray | WYA | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.49% | 17.28% | 10.30% | 3-{[methyl(pyrido[2,3-b]pyrazin-6-yl)amino]methyl}[1,2,4]triazolo[4,3-a]pyrazin-8(7H)-one | 3-{[methyl(pyrido[2,3-b]pyrazin-6-yl)amino]methyl}[1,2,4]triazolo[4,3-a]pyrazin-8(7H)-one | XDS | 100.00% | 6.77 | NSLS-II (17-ID-2) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 68.7 | - | No | - | 94.4 | 52.3 | 66.0 | - | ||||||
| 5sp2 | 0.97 Å | 2022-07-13 | X-ray | S3E, WYG | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.83% | 15.45% | 6.30% | [(2R)-4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)morpholin-2-yl]acetic acid; [(2S)-4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)morpholin-2-yl]acetic acid | [(2S)-4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)morpholin-2-yl]acetic acid; [(2R)-4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)morpholin-2-yl]acetic acid | XDS | 100.00% | 11.44 | NSLS-II (17-ID-2) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 66.6 | - | No | - | 97.8 | 38.5 | 72.0 | - | ||||||
| 5sp3 | 1.01 Å | 2022-07-13 | X-ray | WYJ | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.11% | 15.98% | 7.40% | [(2S,6R)-6-methyl-4-(9H-pyrimido[4,5-b]indol-4-yl)morpholin-2-yl]methanol | [(2S,6R)-6-methyl-4-(9H-pyrimido[4,5-b]indol-4-yl)morpholin-2-yl]methanol | XDS | 99.60% | 10.14 | NSLS-II (17-ID-2) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 67.9 | - | No | - | 97.1 | 27.9 | 86.2 | - | ||||||
| 5sp4 | 1.06 Å | 2022-07-13 | X-ray | RWL | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.81% | 17.76% | 11.30% | 9-[(2-chloro-1,3-thiazol-4-yl)methyl]-9H-purine-2,6-diamine | 9-[(2-chloro-1,3-thiazol-4-yl)methyl]-9H-purine-2,6-diamine | XDS | 99.80% | 6.87 | SSRL (BL12-1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 49.1 | - | No | - | 93.0 | 11.5 | 68.9 | - | ||||||
| 5sp6 | 1.07 Å | 2022-07-13 | X-ray | RXX | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 17.25% | 20.42% | 10.70% | 9-{[(2P)-2-(5-methylfuran-2-yl)-1,3-thiazol-4-yl]methyl}-9H-purine-2,6-diamine | 9-{[(2P)-2-(5-methylfuran-2-yl)-1,3-thiazol-4-yl]methyl}-9H-purine-2,6-diamine | XDS | 99.40% | 6.81 | SSRL (BL12-2) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 40.1 | - | No | - | 78.7 | 14.1 | 63.3 | - | ||||||
| 5sp7 | 1.05 Å | 2022-07-13 | X-ray | RVS | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.38% | 15.18% | 3.60% | (1S,2S)-1-[4-(cyclopropylcarbamamido)benzamido]-1,2,3,4-tetrahydronaphthalene-2-carboxylic acid | (1S,2S)-1-[4-(cyclopropylcarbamamido)benzamido]-1,2,3,4-tetrahydronaphthalene-2-carboxylic acid | XDS | 99.70% | 23.08 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 77.3 | - | No | - | 98.1 | 59.3 | 76.8 | - | ||||||
| 5sp8 | 1.05 Å | 2022-07-13 | X-ray | RXI | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.44% | 16.19% | 4.50% | (6~{S})-7-[4-(cyclopropylcarbamoylamino)phenyl]carbonyl-3-methyl-6,8-dihydro-5~{H}-[1,2,4]triazolo[4,3-a]pyrazine-6-carboxylic acid | (6~{S})-7-[4-(cyclopropylcarbamoylamino)phenyl]carbonyl-3-methyl-6,8-dihydro-5~{H}-[1,2,4]triazolo[4,3-a]pyrazine-6-carboxylic acid | XDS | 99.80% | 15.69 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 68.5 | - | No | - | 96.8 | 41.0 | 74.6 | - | ||||||
| 5sp9 | 1.05 Å | 2022-07-13 | X-ray | RWC | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.01% | 14.59% | 4.70% | (3S)-1-[4-(cyclopropylcarbamamido)benzoyl]-1,2,3,4-tetrahydroquinoline-3-carboxylic acid | (3S)-1-[4-(cyclopropylcarbamamido)benzoyl]-1,2,3,4-tetrahydroquinoline-3-carboxylic acid | XDS | 99.90% | 15.99 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 82.0 | - | No | - | 98.7 | 67.2 | 84.3 | - | ||||||
| 5spa | 1.05 Å | 2022-07-13 | X-ray | RYC, RY6 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.72% | 15.50% | 5.10% | (1S,2S)-1-(4-carbamamidobenzamido)-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid; (1R,2R)-1-(4-carbamamidobenzamido)-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid | (1R,2R)-1-(4-carbamamidobenzamido)-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid; (1S,2S)-1-(4-carbamamidobenzamido)-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.80% | 14.18 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 78.2 | - | No | - | 97.8 | 55.7 | 83.5 | - | ||||||
| 5spb | 1.05 Å | 2022-07-13 | X-ray | S09, S0T | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.62% | 16.52% | 8.10% | (1R,2R)-4-hydroxy-1-[4-(phenylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid; (1S,2S)-4-hydroxy-1-[4-(phenylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid | (1R,2R)-4-hydroxy-1-[4-(phenylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid; (1S,2S)-4-hydroxy-1-[4-(phenylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.70% | 8.55 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 74.6 | - | No | - | 96.2 | 59.3 | 71.4 | - | ||||||
| 5spc | 1.05 Å | 2022-07-13 | X-ray | QLF, QLU | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.46% | 15.33% | 5.30% | (1R,2R)-4-hydroxy-1-[(5,6,7,8-tetrahydro-1,8-naphthyridine-3-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid; (1S,2S)-4-hydroxy-1-[(5,6,7,8-tetrahydro-1,8-naphthyridine-3-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid | (1S,2S)-4-hydroxy-1-[(5,6,7,8-tetrahydro-1,8-naphthyridine-3-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid; (1R,2R)-4-hydroxy-1-[(5,6,7,8-tetrahydro-1,8-naphthyridine-3-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.70% | 14.57 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 74.6 | - | No | - | 97.9 | 59.3 | 69.6 | - | ||||||
| 5spd | 1.05 Å | 2022-07-13 | X-ray | QRU, QYJ | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.32% | 16.23% | 6.00% | (1S,2S)-1-[4-(cyclopropylcarbamamido)benzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid; (1R,2R)-1-[4-(cyclopropylcarbamamido)benzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid | (1R,2R)-1-[4-(cyclopropylcarbamamido)benzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid; (1S,2S)-1-[4-(cyclopropylcarbamamido)benzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.60% | 11.9 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 70.8 | - | No | - | 96.7 | 46.2 | 74.5 | - | ||||||
| 5spe | 1.05 Å | 2022-07-13 | X-ray | S1F | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 12.94% | 14.40% | 3.30% | (1S,2S)-1-{4-[(methoxycarbonyl)amino]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid | (1S,2S)-1-{4-[(methoxycarbonyl)amino]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.90% | 23.69 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 79.4 | - | No | - | 98.8 | 59.3 | 82.4 | - | ||||||
| 5spf | 1.05 Å | 2022-07-13 | X-ray | RYQ | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.23% | 15.98% | 3.30% | 9-[(2-methyl-1,3-thiazol-4-yl)methyl]-9H-purine-2,6-diamine | 9-[(2-methyl-1,3-thiazol-4-yl)methyl]-9H-purine-2,6-diamine | XDS | 99.70% | 24.23 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 65.6 | - | No | - | 97.1 | 38.5 | 70.5 | - | ||||||
| 5spg | 1.05 Å | 2022-07-13 | X-ray | RZI | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.77% | 15.79% | 5.10% | 9-[(2-cyclopropyl-1,3-thiazol-4-yl)methyl]-9H-purine-2,6-diamine | 9-[(2-cyclopropyl-1,3-thiazol-4-yl)methyl]-9H-purine-2,6-diamine | XDS | 99.90% | 14.29 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 58.3 | - | No | - | 97.4 | 20.8 | 72.9 | - | ||||||
| 5sph | 1.05 Å | 2022-07-13 | X-ray | RZR | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.35% | 15.08% | 4.60% | (1R,2S)-1-[4-(methylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid | (1R,2S)-1-[4-(methylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.80% | 15.73 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 79.9 | - | No | - | 98.2 | 63.1 | 81.1 | - | ||||||
| 5spi | 1.05 Å | 2022-07-13 | X-ray | S1X, S2R | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 12.84% | 14.51% | 3.50% | (1S,2S)-1-{4-[(cyclopropanecarbonyl)amino]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid; (1R,2R)-1-{4-[(cyclopropanecarbonyl)amino]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid | (1S,2S)-1-{4-[(cyclopropanecarbonyl)amino]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid; (1R,2R)-1-{4-[(cyclopropanecarbonyl)amino]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.90% | 22.33 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 80.8 | - | No | - | 98.7 | 63.1 | 83.7 | - | ||||||
| 5spj | 1.05 Å | 2022-07-13 | X-ray | S3R | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 12.91% | 14.63% | 4.50% | 5-chloro-N~3~-[(4-cyclopropyl-5-methyl-4H-1,2,4-triazol-3-yl)methyl]pyrazine-2,3-diamine | 5-chloro-N~3~-[(4-cyclopropyl-5-methyl-4H-1,2,4-triazol-3-yl)methyl]pyrazine-2,3-diamine | XDS | 99.80% | 16.87 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 81.0 | - | No | - | 98.6 | 67.2 | 80.3 | - | ||||||
| 5spk | 1.05 Å | 2022-07-13 | X-ray | S4F | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.13% | 15.97% | 4.70% | 3-[(3R)-1-(6-amino-5-chloropyrimidin-4-yl)piperidin-3-yl]propanoic acid | 3-[(3R)-1-(6-amino-5-chloropyrimidin-4-yl)piperidin-3-yl]propanoic acid | XDS | 99.80% | 15.12 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 72.2 | - | No | - | 97.1 | 46.2 | 77.4 | - | ||||||
| 5spl | 1.05 Å | 2022-07-13 | X-ray | RW5 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 12.89% | 14.44% | 4.30% | [(2S,4S)-4-methyl-2-(5-methylfuran-2-yl)piperidin-1-yl](7H-pyrrolo[2,3-d]pyrimidin-4-yl)methanone | [(2S,4S)-4-methyl-2-(5-methylfuran-2-yl)piperidin-1-yl](7H-pyrrolo[2,3-d]pyrimidin-4-yl)methanone | XDS | 99.80% | 18.11 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 69.1 | - | No | - | 98.8 | 55.7 | 59.3 | - | ||||||
| 5spm | 1.05 Å | 2022-07-13 | X-ray | S4O | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 12.82% | 14.57% | 4.20% | 4-hydroxy-6-(3-hydroxy-1-methyl-1,4,5,7-tetrahydro-6H-pyrazolo[3,4-c]pyridine-6-carbonyl)-2H-pyran-2-one | 4-hydroxy-6-(3-hydroxy-1-methyl-1,4,5,7-tetrahydro-6H-pyrazolo[3,4-c]pyridine-6-carbonyl)-2H-pyran-2-one | XDS | 99.70% | 18.11 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 73.9 | - | No | - | 98.7 | 46.2 | 80.1 | - | ||||||
| 5spn | 1.05 Å | 2022-07-13 | X-ray | S50 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.75% | 15.52% | 4.50% | 1-cyclopentyl-3-methyl-N-(1H-pyrrolo[2,3-b]pyridin-5-yl)-1H-pyrazole-5-sulfonamide | 1-cyclopentyl-3-methyl-N-(1H-pyrrolo[2,3-b]pyridin-5-yl)-1H-pyrazole-5-sulfonamide | XDS | 99.70% | 15.59 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 67.9 | - | No | - | 97.7 | 46.2 | 67.2 | - | ||||||
| 5spo | 1.05 Å | 2022-07-13 | X-ray | S5F | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.47% | 15.20% | 3.20% | 6-chloro-4-{(8S)-8-[(4H-1,2,4-triazol-4-yl)methyl]-6-azaspiro[3.4]octan-6-yl}pyrimidin-2(1H)-one | 6-chloro-4-{(8S)-8-[(4H-1,2,4-triazol-4-yl)methyl]-6-azaspiro[3.4]octan-6-yl}pyrimidin-2(1H)-one | XDS | 99.80% | 24.89 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 81.6 | - | No | - | 98.1 | 67.2 | 83.1 | - | ||||||
| 5spp | 1.05 Å | 2022-07-13 | X-ray | S5U | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.72% | 15.56% | 3.90% | 4-[methyl(7H-pyrrolo[2,3-d]pyrimidine-4-carbonyl)amino]butanoic acid | 4-[methyl(7H-pyrrolo[2,3-d]pyrimidine-4-carbonyl)amino]butanoic acid | XDS | 99.90% | 18.52 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 70.3 | - | No | - | 97.7 | 59.3 | 59.3 | - | ||||||
| 5spq | 1.05 Å | 2022-07-13 | X-ray | S63 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.58% | 16.36% | 5.50% | (3R)-1-[(4-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)acetyl]-3-methylpyrrolidine-3-carboxylic acid | (3R)-1-[(4-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)acetyl]-3-methylpyrrolidine-3-carboxylic acid | XDS | 99.80% | 12.29 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 75.1 | - | No | - | 96.5 | 63.1 | 68.6 | - | ||||||
| 5spr | 1.05 Å | 2022-07-13 | X-ray | S6C | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.08% | 15.87% | 4.10% | [(3S)-1-(7H-purin-6-yl)piperidin-3-yl]acetic acid | [(3S)-1-(7H-purin-6-yl)piperidin-3-yl]acetic acid | XDS | 99.80% | 17.52 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 62.3 | - | No | - | 97.3 | 27.9 | 74.0 | - | ||||||
| 5sps | 1.05 Å | 2022-07-13 | X-ray | S6N | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.98% | 16.70% | 3.60% | (3S)-6,6-dimethyl-1-(7H-pyrrolo[2,3-d]pyrimidine-4-carbonyl)piperidine-3-carboxylic acid | (3S)-6,6-dimethyl-1-(7H-pyrrolo[2,3-d]pyrimidine-4-carbonyl)piperidine-3-carboxylic acid | XDS | 99.90% | 19.95 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 57.3 | - | No | - | 95.8 | 34.4 | 58.9 | - | ||||||
| 5spt | 1.05 Å | 2022-07-13 | X-ray | S6U | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.18% | 16.07% | 4.90% | 2-(2-oxo-1,3-oxazolidin-3-yl)ethyl 7H-pyrrolo[2,3-d]pyrimidine-4-carboxylate | 2-(2-oxo-1,3-oxazolidin-3-yl)ethyl 7H-pyrrolo[2,3-d]pyrimidine-4-carboxylate | XDS | 99.80% | 14.77 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 71.5 | - | No | - | 97.0 | 49.1 | 73.1 | - | ||||||
| 5spu | 1.05 Å | 2022-07-13 | X-ray | S7F | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.76% | 15.47% | 4.50% | (3S)-1-(6-amino-5-methylpyridine-3-sulfonyl)piperidine-3-carboxamide | (3S)-1-(6-amino-5-methylpyridine-3-sulfonyl)piperidine-3-carboxamide | XDS | 99.90% | 16.44 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 80.4 | - | No | - | 97.8 | 67.2 | 79.3 | - | ||||||
| 5spv | 1.05 Å | 2022-07-13 | X-ray | S7O | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.80% | 16.74% | 8.30% | 2-[methyl-[(9-oxidanylidene-1$l^{4},7,8-triazabicyclo[4.3.0]nona-1(6),2,4-trien-3-yl)carbonyl]amino]ethanoic acid | 2-[methyl-[(9-oxidanylidene-1$l^{4},7,8-triazabicyclo[4.3.0]nona-1(6),2,4-trien-3-yl)carbonyl]amino]ethanoic acid | XDS | 99.90% | 7.99 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 74.5 | - | No | - | 95.7 | 63.1 | 67.7 | - | ||||||
| 5spw | 1.05 Å | 2022-07-13 | X-ray | S5O | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.60% | 15.25% | 3.80% | (1R,5S,6R)-3-(7H-purin-6-yl)-3-azabicyclo[3.2.2]nonane-6-carboxylic acid | (1R,5S,6R)-3-(7H-purin-6-yl)-3-azabicyclo[3.2.2]nonane-6-carboxylic acid | XDS | 99.80% | 19.65 | ALS (8.3.1) | PHENIX | 2022-06-09 | 36598939 | - | - | - | - | N/A | - | - | Non-structural protein 3 | 68.7 | - | No | - | 98.0 | 46.2 | 68.6 | - | ||||||
| 7fb0 | 3.40 Å | 2022-07-13 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2021-07-08 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7fb1 | 3.70 Å | 2022-07-13 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-07-08 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7fb3 | 3.10 Å | 2022-07-13 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-07-08 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7fb4 | 3.40 Å | 2022-07-13 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-07-08 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7fbj | 2.85 Å | 2022-07-13 | X-ray | NAG | SARS-CoV-2 | Spike | Functional ligand | 25.37% | 30.04% | 12.90% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | DIALS | 99.80% | 7.2 | SSRF (BL18U1) | REFMAC | 2021-07-11 | 35583124 | - | - | - | - | N/A | - | - | New antigen receptor variable domain; Spike protein S1 - receptor binding domain | 1.6 | - | No | - | 7.2 | 16.5 | 27.0 | - | ||||||
| 7fbk | 1.90 Å | 2022-07-13 | X-ray | - | SARS-CoV-2 | Spike | No functional ligands | 19.69% | 23.94% | 7.30% | - | XDS | 99.90% | 16.5 | SSRF (BL18U1) | REFMAC | 2021-07-11 | 35583124 | - | - | - | - | N/A | - | - | New antigen receptor variable domain; Spike protein S1 - receptor binding domain | 17.7 | - | No | - | 45.4 | 18.8 | 45.0 | - | |||||||
| 7fc6 | 2.65 Å | 2022-07-13 | X-ray | NAG | SARS-CoV | Spike | PPI | 18.49% | 23.49% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.30% | 16.7 | SSRF (BL17U1) | PHENIX | 2021-07-13 | 35917815 | - | - | - | - | N/A | - | - | Spike protein S1; Angiotensin-converting enzyme | 40.3 | - | No | - | 50.2 | 81.3 | 24.9 | - | ||||||
| 7p19 | 3.24 Å | 2022-07-13 | X-ray | NAG | SARS-CoV-2 | Spike | PPI | 23.09% | 28.85% | 25.60% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | autoPROC | 99.43% | 7.4 | ALBA (XALOC) | PHENIX | 2021-07-01 | 36048155 | ZN | - | - | - | N/A | - | - | Spike protein S1; Processed angiotensin-converting enzyme 2 | 29.0 | - | No | - | 10.5 | 58.9 | 64.4 | - | ||||||
| 7wjy | 3.24 Å | 2022-07-13 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-08 | 35768499 | - | - | - | - | N/A | - | - | 6m6 light chain; 6m6 heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7wjz | 3.34 Å | 2022-07-13 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-08 | 35768499 | - | - | - | - | N/A | - | - | 6m6 light chain; 6m6 heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7wk0 | 3.32 Å | 2022-07-13 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-08 | 35768499 | - | - | - | - | N/A | - | - | Spike protein S1 - RBD; 6m6 heavy chain; 6m6 light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7won | 3.90 Å | 2022-07-13 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-01-22 | 35260792 | - | - | - | - | N/A | - | - | Spike protein S1; VacW-209 heavy chain; VacW-209 light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wp1 | 3.77 Å | 2022-07-13 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-01-22 | 35260792 | - | - | - | - | N/A | - | - | Spike protein S1; VacW-209 light chain; VacW-209 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wp5 | 3.90 Å | 2022-07-13 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-01-22 | 35260792 | - | - | - | - | N/A | - | - | VacW-209 light chain; Spikeprotein S1; VacW-209 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7x6a | 3.50 Å | 2022-07-13 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-03-07 | 35714668 | - | - | - | - | N/A | - | - | Spike glycoprotein; Heavy chain of Fab BD55-5840; Light chain of Fab BD55-5840 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7xiw | 3.62 Å | 2022-07-13 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-04-14 | 35714668 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7xix | 3.25 Å | 2022-07-13 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-04-14 | 35714668 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7xiy | 3.07 Å | 2022-07-13 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-04-14 | 35714668 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7xiz | 3.74 Å | 2022-07-13 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-04-14 | 35714668 | - | - | - | - | N/A | - | - | Spike protein S1 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7xnq | 3.52 Å | 2022-07-13 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-04-29 | 35714668 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7xnr | 3.49 Å | 2022-07-13 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-04-29 | 35714668 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7xns | 3.48 Å | 2022-07-13 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-04-29 | 35714668 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7z6v | 3.10 Å | 2022-07-13 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-03-14 | 35858383 | - | - | - | - | N/A | - | - | Nanobody H11; Spike glycoprotein,Fibritin | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7z7x | 3.30 Å | 2022-07-13 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-03-16 | 35858383 | - | - | - | - | N/A | - | - | Spike glycoprotein,Fibritin; Nanobody H11-H6 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7z85 | 3.10 Å | 2022-07-13 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-03-16 | 35858383 | - | - | - | - | N/A | - | - | Nanobody H11-B5; Spike glycoprotein,Fibritin | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7z86 | 3.40 Å | 2022-07-13 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-03-16 | 35858383 | - | - | - | - | N/A | - | - | Nanobody H11-H4 Q98R H100E; Spike glycoprotein,Fibritin | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7z9q | 3.60 Å | 2022-07-13 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | 31.25% | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | REFMAC | 2022-03-21 | 35858383 | - | - | - | - | N/A | - | - | Nanobody H11-A10; Spike glycoprotein,Fibritin | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7z9r | 4.20 Å | 2022-07-13 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-03-21 | 35858383 | - | - | - | - | N/A | - | - | Nanobody H11-H4 Q98R H100E; Spike glycoprotein,Fibritin | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8d4j | 1.78 Å | 2022-07-13 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 16.56% | 20.92% | 6.30% | - | HKL-2000 | 99.50% | 24.2 | APS (22-BM) | REFMAC | 2022-06-02 | 36119652 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 69.9 | - | No | - | 74.7 | 53.7 | 87.0 | - | |||||||
| 8d4k | 1.89 Å | 2022-07-13 | X-ray | K36 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.56% | 22.36% | 8.40% | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | HKL-2000 | 98.60% | 20.35 | APS (22-ID) | REFMAC | 2022-06-02 | 36119652 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 74.2 | - | No | - | 61.6 | 88.6 | 75.6 | - | ||||||
| 8d4l | 1.70 Å | 2022-07-13 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 17.10% | 21.28% | 5.50% | - | HKL-2000 | 96.69% | 28.72 | APS (22-ID) | REFMAC | 2022-06-02 | 36119652 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 51.3 | - | No | - | 71.8 | 39.9 | 65.9 | - | |||||||
| 8d4m | 1.81 Å | 2022-07-13 | X-ray | K36 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 17.17% | 22.89% | 7.40% | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | HKL-2000 | 98.10% | 21.77 | APS (22-ID) | REFMAC | 2022-06-02 | 36119652 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 66.4 | - | No | - | 56.3 | 78.7 | 72.9 | - | ||||||
| 8d4n | 2.70 Å | 2022-07-13 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 20.62% | 25.82% | 17.20% | - | HKL-2000 | 99.10% | 9.13 | APS (22-BM) | REFMAC | 2022-06-02 | 36119652 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 56.1 | - | No | - | 28.0 | 73.6 | 85.1 | - | |||||||
| 8dcz | 2.38 Å | 2022-07-13 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.19% | 25.13% | 9.20% | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | HKL-2000 | 98.60% | 12.33 | APS (22-ID) | REFMAC | 2022-06-17 | 37637734 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 45.2 | - | No | - | 34.0 | 71.3 | 60.5 | - | ||||||
| 8dd1 | 2.03 Å | 2022-07-13 | X-ray | K36 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.32% | 24.99% | 7.80% | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | HKL-2000 | 97.00% | 22.18 | APS (22-ID) | REFMAC | 2022-06-17 | 37637734 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 61.5 | - | No | - | 35.4 | 73.6 | 88.6 | - | ||||||
| 8dd9 | 2.04 Å | 2022-07-13 | X-ray | B1S | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 16.52% | 22.84% | 9.70% | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | HKL-2000 | 96.90% | 13.59 | APS (22-ID) | REFMAC | 2022-06-17 | 37637734 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 62.7 | - | No | - | 56.6 | 68.7 | 74.7 | - | ||||||
| 8dfe | 1.89 Å | 2022-07-13 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 20.84% | 26.77% | 7.10% | - | HKL-2000 | 94.00% | 26.7 | APS (22-ID) | REFMAC | 2022-06-22 | 37637734 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 48.1 | - | No | - | 21.1 | 59.6 | 90.7 | - | |||||||
| 8dfn | 2.04 Å | 2022-07-13 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | No functional ligands | 19.09% | 25.08% | 8.70% | - | HKL-2000 | 98.60% | 16.75 | APS (22-ID) | REFMAC | 2022-06-22 | 37637734 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 61.7 | - | No | - | 34.6 | 71.2 | 92.2 | - | |||||||
| 8dgb | 2.87 Å | 2022-07-13 | X-ray | B1S | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 22.71% | 27.22% | 7.90% | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | HKL-2000 | 93.50% | 9.03 | APS (22-BM) | REFMAC | 2022-06-23 | 37637734 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 56.4 | - | No | - | 18.3 | 93.3 | 75.9 | - | ||||||
| 7en8 | 1.83 Å | 2022-07-20 | X-ray | J7R | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 22.48% | 27.37% | 12.50% | ~{N}-[(1~{S},2~{R})-2-[[4-bromanyl-2-(methylcarbamoyl)-6-nitro-phenyl]amino]cyclohexyl]isoquinoline-4-carboxamide | ~{N}-[(1~{S},2~{R})-2-[[4-bromanyl-2-(methylcarbamoyl)-6-nitro-phenyl]amino]cyclohexyl]isoquinoline-4-carboxamide | CrysalisPro | 99.27% | 21.7 | ROTATING ANODE () | PHENIX | 2021-04-16 | 36844503 | - | - | - | - | N/A | - | - | 3C-like proteinase | 27.8 | - | No | - | 17.5 | 41.1 | 72.8 | - | ||||||
| 7en9 | 1.90 Å | 2022-07-20 | X-ray | J7O | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.52% | 23.16% | 13.39% | 5-bromanyl-~{N}-methyl-3-nitro-2-[(4~{R},5~{S})-2-(7-oxidanylisoquinolin-4-yl)carbonyl-4-phenyl-2,7-diazaspiro[4.4]nonan-7-yl]benzamide | 5-bromanyl-~{N}-methyl-3-nitro-2-[(4~{R},5~{S})-2-(7-oxidanylisoquinolin-4-yl)carbonyl-4-phenyl-2,7-diazaspiro[4.4]nonan-7-yl]benzamide | CrysalisPro | 99.61% | 18.24 | ROTATING ANODE () | PHENIX | 2021-04-16 | 36844503 | - | - | - | - | N/A | - | - | 3C-like proteinase | 47.6 | - | No | - | 53.5 | 42.8 | 74.2 | - | ||||||
| 7end | 1.99 Å | 2022-07-20 | X-ray | J7R | SARS-CoV | NSP5 (3CLpro) | Functional ligand | 19.74% | 23.70% | 12.36% | ~{N}-[(1~{S},2~{R})-2-[[4-bromanyl-2-(methylcarbamoyl)-6-nitro-phenyl]amino]cyclohexyl]isoquinoline-4-carboxamide | ~{N}-[(1~{S},2~{R})-2-[[4-bromanyl-2-(methylcarbamoyl)-6-nitro-phenyl]amino]cyclohexyl]isoquinoline-4-carboxamide | CrysalisPro | 99.46% | 33.16 | ROTATING ANODE () | PHENIX | 2021-04-16 | 36844503 | - | - | - | - | N/A | - | - | Replicase polyprotein 1a | 66.4 | - | No | - | 47.7 | 78.7 | 81.5 | - | ||||||
| 7ene | 2.98 Å | 2022-07-20 | X-ray | J7R | MERS-CoV | NSP5 (3CLpro) | Functional ligand | 23.49% | 27.45% | N/A | ~{N}-[(1~{S},2~{R})-2-[[4-bromanyl-2-(methylcarbamoyl)-6-nitro-phenyl]amino]cyclohexyl]isoquinoline-4-carboxamide | ~{N}-[(1~{S},2~{R})-2-[[4-bromanyl-2-(methylcarbamoyl)-6-nitro-phenyl]amino]cyclohexyl]isoquinoline-4-carboxamide | CrysalisPro | 98.42% | 10.58 | ROTATING ANODE () | PHENIX | 2021-04-16 | 36844503 | - | - | - | - | N/A | - | - | ORF1a protein - UNP residues 3248-3553 | 36.5 | - | No | - | 17.0 | 76.2 | 56.0 | - | ||||||
| 7p2o | - | 2022-07-20 | NMR | - | SARS-CoV-2 | NSP3(Cleavage_domain) | No functional ligands | N/A | N/A | N/A | - | - | - | () | 2021-07-06 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7rvm | 1.95 Å | 2022-07-20 | X-ray | 7V2 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 20.55% | 23.83% | N/A | N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | iMOSFLM | 97.40% | 9.3 | ROTATING ANODE () | PHENIX | 2021-08-19 | 35779291 | - | - | - | - | N/A | - | - | 3C-like proteinase | 23.7 | - | No | - | 46.5 | 28.0 | 48.2 | - | ||||||
| 7rvn | 1.63 Å | 2022-07-20 | X-ray | 7VB | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.72% | 21.24% | N/A | N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methylidene-L-norvalinamide | N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methylidene-L-norvalinamide | iMOSFLM | 95.80% | 12.5 | ROTATING ANODE () | PHENIX | 2021-08-19 | 35779291 | - | - | - | - | N/A | - | - | 3C-like proteinase | 34.4 | - | No | - | 72.0 | 19.5 | 53.4 | - | ||||||
| 7rvo | 1.80 Å | 2022-07-20 | X-ray | 7VI | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 24.44% | 27.50% | N/A | N-[(benzyloxy)carbonyl]-L-valyl-3-cyclopropyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | N-[(benzyloxy)carbonyl]-L-valyl-3-cyclopropyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | iMOSFLM | 89.80% | 3.5 | ALS (5.0.2) | PHENIX | 2021-08-19 | 35779291 | - | - | - | - | N/A | - | - | 3C-like proteinase | 2.0 | - | No | - | 16.6 | 11.4 | 25.4 | - | ||||||
| 7rvp | 1.90 Å | 2022-07-20 | X-ray | 7VQ | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 23.68% | 27.36% | N/A | N-[(benzyloxy)carbonyl]-L-valyl-3-furan-2-yl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | N-[(benzyloxy)carbonyl]-L-valyl-3-furan-2-yl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | iMOSFLM | 95.50% | 5.9 | ALS (5.0.2) | PHENIX | 2021-08-19 | 35779291 | - | - | - | - | N/A | - | - | 3C-like proteinase | 7.0 | - | No | - | 17.5 | 23.6 | 36.9 | - | ||||||
| 7rvq | 2.48 Å | 2022-07-20 | X-ray | 7VW | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 23.86% | 32.22% | N/A | N-[(benzyloxy)carbonyl]-O-tert-butyl-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | N-[(benzyloxy)carbonyl]-O-tert-butyl-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | PROTEUM PLUS | 98.10% | 3.5 | ROTATING ANODE () | PHENIX | 2021-08-19 | 35779291 | - | - | - | - | N/A | - | - | 3C-like proteinase | 6.2 | - | No | - | 4.8 | 31.1 | 39.0 | - | ||||||
| 7rvr | 2.46 Å | 2022-07-20 | X-ray | 7W5 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 25.29% | 32.31% | N/A | N-[(benzyloxy)carbonyl]-3-methyl-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | N-[(benzyloxy)carbonyl]-3-methyl-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | PROTEUM PLUS | 98.40% | 8.0 | ROTATING ANODE () | PHENIX | 2021-08-19 | 35779291 | - | - | - | - | N/A | - | - | 3C-like proteinase | 3.8 | - | No | - | 4.8 | 18.9 | 40.4 | - | ||||||
| 7rvs | 1.85 Å | 2022-07-20 | X-ray | 81L | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.93% | 22.65% | N/A | N-[(benzyloxy)carbonyl]-3-methyl-L-valyl-3-cyclopropyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | N-[(benzyloxy)carbonyl]-3-methyl-L-valyl-3-cyclopropyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | PROTEUM PLUS | 99.90% | 17.8 | ROTATING ANODE () | PHENIX | 2021-08-19 | 35779291 | - | - | - | - | N/A | - | - | 3C-like proteinase | 27.5 | - | No | - | 58.7 | 13.8 | 58.0 | - | ||||||
| 7rvt | 2.10 Å | 2022-07-20 | X-ray | 7XK | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 27.02% | 30.76% | N/A | N~2~-[(2S)-2-{[(benzyloxy)carbonyl]amino}-2-cyclopropylacetyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | N~2~-[(2S)-2-{[(benzyloxy)carbonyl]amino}-2-cyclopropylacetyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | iMOSFLM | 95.80% | 3.8 | ALS (5.0.2) | PHENIX | 2021-08-19 | 35779291 | - | - | - | - | N/A | - | - | 3C-like proteinase | 1.2 | - | No | - | 6.2 | 11.8 | 28.9 | - | ||||||
| 7rvu | 2.50 Å | 2022-07-20 | X-ray | 7XT | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 25.15% | 32.85% | N/A | N-[(benzyloxy)carbonyl]-3-methyl-L-isovalyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | N-[(benzyloxy)carbonyl]-3-methyl-L-isovalyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | iMOSFLM | 98.90% | 2.1 | ALS (5.0.2) | PHENIX | 2021-08-19 | 35779291 | - | - | - | - | N/A | - | - | 3C-like proteinase | 2.1 | - | No | - | 4.5 | 25.0 | 24.6 | - | ||||||
| 7rvv | 3.00 Å | 2022-07-20 | X-ray | 7Y2 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 34.14% | 42.79% | N/A | N-[(benzyloxy)carbonyl]-2-methyl-L-alanyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | N-[(benzyloxy)carbonyl]-2-methyl-L-alanyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | PROTEUM PLUS | 93.20% | 6.1 | ROTATING ANODE () | PHENIX | 2021-08-19 | 35779291 | - | - | - | - | N/A | - | - | 3C-like proteinase | 0.8 | - | No | - | 3.4 | 23.9 | 15.4 | - | ||||||
| 7rvw | 1.85 Å | 2022-07-20 | X-ray | 7YB | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 18.98% | 21.75% | N/A | benzyl (1-{[(2S)-3-cyclohexyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamoyl}cyclopropyl)carbamate | benzyl (1-{[(2S)-3-cyclohexyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamoyl}cyclopropyl)carbamate | PROTEUM PLUS | 99.90% | 13.3 | ROTATING ANODE () | PHENIX | 2021-08-19 | 35779291 | - | - | - | - | N/A | - | - | 3C-like proteinase | 37.6 | - | No | - | 67.5 | 22.0 | 61.7 | - | ||||||
| 7rvx | 1.85 Å | 2022-07-20 | X-ray | 7YI | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 23.53% | 26.51% | N/A | benzyl [(1S)-1-cyclopropyl-2-{[(2S)-3-cyclopropyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]amino}-2-oxoethyl]carbamate | benzyl [(1S)-1-cyclopropyl-2-{[(2S)-3-cyclopropyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]amino}-2-oxoethyl]carbamate | iMOSFLM | 89.10% | 7.6 | ROTATING ANODE () | PHENIX | 2021-08-19 | 35779291 | - | - | - | - | N/A | - | - | 3C-like proteinase | 7.4 | - | No | - | 22.8 | 17.1 | 39.5 | - | ||||||
| 7rvy | 1.85 Å | 2022-07-20 | X-ray | 7YQ | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 23.09% | 26.80% | N/A | O-tert-butyl-N-{[(3-chlorophenyl)methoxy]carbonyl}-L-threonyl-3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | O-tert-butyl-N-{[(3-chlorophenyl)methoxy]carbonyl}-L-threonyl-3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | PROTEUM PLUS | 92.60% | 12.2 | ROTATING ANODE () | PHENIX | 2021-08-19 | 35779291 | - | - | - | - | N/A | - | - | 3C-like proteinase | 7.1 | - | No | - | 20.8 | 19.6 | 38.0 | - | ||||||
| 7rvz | 1.90 Å | 2022-07-20 | X-ray | 7YW | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 19.11% | 21.99% | N/A | O-tert-butyl-N-{[(3-chlorophenyl)methoxy]carbonyl}-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | O-tert-butyl-N-{[(3-chlorophenyl)methoxy]carbonyl}-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | iMOSFLM | 96.10% | 8.7 | ALS (5.0.2) | PHENIX | 2021-08-19 | 35779291 | - | - | - | - | N/A | - | - | 3C-like proteinase | 37.0 | - | No | - | 65.3 | 26.0 | 58.8 | - | ||||||
| 7rw0 | 1.85 Å | 2022-07-20 | X-ray | 7YZ | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 22.20% | 25.34% | N/A | N-{[(3-chlorophenyl)methoxy]carbonyl}-L-valyl-3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | N-{[(3-chlorophenyl)methoxy]carbonyl}-L-valyl-3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | PROTEUM PLUS | 99.80% | 8.8 | ROTATING ANODE () | PHENIX | 2021-08-19 | 35779291 | - | - | - | - | N/A | - | - | 3C-like proteinase | 11.4 | - | No | - | 32.2 | 18.9 | 41.7 | - | ||||||
| 7rw1 | 2.50 Å | 2022-07-20 | X-ray | 800 | SARS-CoV-2 | NSP5 (3CLpro) | Functional ligand | 25.66% | 31.60% | N/A | N-(1H-indole-2-carbonyl)-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | N-(1H-indole-2-carbonyl)-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | iMOSFLM | 97.10% | 4.7 | ALS (5.0.2) | PHENIX | 2021-08-19 | 35779291 | - | - | - | - | N/A | - | - | 3C-like proteinase | 15.2 | - | No | - | 5.3 | 73.4 | 24.4 | - | ||||||
| 7vq0 | 3.03 Å | 2022-07-20 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-10-18 | 35794202 | - | - | - | - | N/A | - | - | Spike glycoprotein; Neutralizing nanobody P86 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7wo4 | 4.47 Å | 2022-07-20 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-20 | 35924918 | - | - | - | - | N/A | - | - | Spike glycoprotein; mAb15 VH; mAb15 VL | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7wo5 | 3.45 Å | 2022-07-20 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-20 | 35924918 | - | - | - | - | N/A | - | - | mAb15 VL; Spike glycoprotein; mAb15 VH | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7wo7 | 3.80 Å | 2022-07-20 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-20 | 35924918 | - | - | - | - | N/A | - | - | mAb15 VL; mAb15 VH; Spike protein S1 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7woa | 3.25 Å | 2022-07-20 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-21 | 35924918 | - | - | - | - | N/A | - | - | mAb60 VH; Spike glycoprotein; mAb60 VL | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7wob | 3.25 Å | 2022-07-20 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-21 | 35924918 | - | - | - | - | N/A | - | - | mAb60 VH; Spike glycoprotein; mAb60 VL | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7woc | 3.35 Å | 2022-07-20 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-21 | 35924918 | - | - | - | - | N/A | - | - | Spike protein S1; mAb60 VH; mAb60 VL | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7wog | 4.06 Å | 2022-07-20 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-01-21 | 35924918 | - | - | - | - | N/A | - | - | 553-49 VL; Spike protein S1 - RBD; 553-49 VH | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wp0 | 3.71 Å | 2022-07-20 | Cryo-EM | NAG | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-22 | 35260792 | - | - | - | - | N/A | - | - | Spike protein S1; IG c335_light_IGLV1-40_IGLJ3; VacW-209 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7wp2 | 3.52 Å | 2022-07-20 | Cryo-EM | - | SARS-CoV-2 | Spike | PPI | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-01-22 | 35260792 | - | - | - | - | N/A | - | - | Spike protein S1; VacW-209 light chain; VacW-209 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wz2 | 2.70 Å | 2022-07-20 | Cryo-EM | NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-16 | 35924918 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7xtz | 2.80 Å | 2022-07-20 | Cryo-EM | NAG, BLA, EIC | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | LINOLEIC ACID; 2-acetamido-2-deoxy-beta-D-glucopyranose; BILIVERDINE IX ALPHA | 2-acetamido-2-deoxy-beta-D-glucopyranose; BILIVERDINE IX ALPHA; LINOLEIC ACID | - | - | () | PHENIX | 2022-05-18 | 35905112 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7xu0 | 2.90 Å | 2022-07-20 | Cryo-EM | NAG, BLA, EIC | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | BILIVERDINE IX ALPHA; LINOLEIC ACID; 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose; BILIVERDINE IX ALPHA; LINOLEIC ACID | - | - | () | PHENIX | 2022-05-18 | 35905112 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7xu1 | 3.00 Å | 2022-07-20 | Cryo-EM | BLA, EIC, NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose; BILIVERDINE IX ALPHA; LINOLEIC ACID | BILIVERDINE IX ALPHA; LINOLEIC ACID; 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-05-18 | 35905112 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7xu2 | 3.20 Å | 2022-07-20 | Cryo-EM | NAG, EIC, BLA | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | BILIVERDINE IX ALPHA; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose; LINOLEIC ACID; BILIVERDINE IX ALPHA | - | - | () | PHENIX | 2022-05-18 | 35905112 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7xu3 | 3.00 Å | 2022-07-20 | Cryo-EM | NAG, BLA | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | BILIVERDINE IX ALPHA | 2-acetamido-2-deoxy-beta-D-glucopyranose; BILIVERDINE IX ALPHA | - | - | () | PHENIX | 2022-05-18 | 35905112 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7xu4 | 3.20 Å | 2022-07-20 | Cryo-EM | EIC, NAG, BLA | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; BILIVERDINE IX ALPHA; LINOLEIC ACID | LINOLEIC ACID; 2-acetamido-2-deoxy-beta-D-glucopyranose; BILIVERDINE IX ALPHA | - | - | () | PHENIX | 2022-05-18 | 35905112 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7xu5 | 3.10 Å | 2022-07-20 | Cryo-EM | NAG, BLA | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | BILIVERDINE IX ALPHA | 2-acetamido-2-deoxy-beta-D-glucopyranose; BILIVERDINE IX ALPHA | - | - | () | PHENIX | 2022-05-18 | 35905112 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 7xu6 | 2.90 Å | 2022-07-20 | Cryo-EM | BLA, NAG | SARS-CoV-2 | Spike | Functional ligand | N/A | N/A | N/A | BILIVERDINE IX ALPHA; 2-acetamido-2-deoxy-beta-D-glucopyranose | BILIVERDINE IX ALPHA; 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-05-18 | 35905112 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||
| 8csa | 3.84 Å | 2022-07-20 | Cryo-EM | - | SARS-CoV-2 | Spike | No functional ligands | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-05-12 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7p5g | - | 2022-07-27 | NMR | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | Spike glycoprotein | - | - | - | () | 2021-07-14 | 36406731 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7p5q | - | 2022-07-27 | NMR | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | Spike glycoprotein | - | - | - | () | 2021-07-14 | 36406731 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7p5s | - | 2022-07-27 | NMR | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | Spike glycoprotein | - | - | - | () | 2021-07-14 | 36406731 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7s6w | 2.29 Å | 2022-07-27 | X-ray | 8G9 | SARS-CoV-2 | NSP5 (3CLpro) | 19.86% | 24.18% | N/A | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | PROTEUM PLUS | 98.70% | 9.8 | ROTATING ANODE () | PHENIX | 2021-09-15 | 35839690 | - | - | - | - | N/A | - | - | 3C-like proteinase | 17.0 | - | No | - | 43.3 | 19.1 | 45.2 | - | |||||||
| 7s6x | 1.80 Å | 2022-07-27 | X-ray | I70 | SARS-CoV-2 | NSP5 (3CLpro) | 18.78% | 20.69% | N/A | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | iMOSFLM | 95.50% | 12.4 | ALS (5.0.2) | PHENIX | 2021-09-15 | 35839690 | - | - | - | - | N/A | - | - | 3C-like proteinase | 40.6 | - | No | - | 76.7 | 36.8 | 43.6 | - | |||||||
| 7s6y | 1.85 Å | 2022-07-27 | X-ray | 8GW | SARS-CoV-2 | NSP5 (3CLpro) | 22.90% | 25.80% | N/A | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-[(cyclopropylmethyl)amino]-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-[(cyclopropylmethyl)amino]-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | PROTEUM PLUS | 99.90% | 12.3 | ROTATING ANODE () | PHENIX | 2021-09-15 | 35839690 | - | - | - | - | N/A | - | - | 3C-like proteinase | 15.1 | - | No | - | 28.1 | 17.2 | 57.6 | - | |||||||
| 7s6z | 1.85 Å | 2022-07-27 | X-ray | I71 | SARS-CoV-2 | NSP5 (3CLpro) | 20.18% | 22.15% | N/A | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-(ethylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-(ethylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | PROTEUM PLUS | 99.90% | 19.4 | ROTATING ANODE () | PHENIX | 2021-09-15 | 35839690 | - | - | - | - | N/A | - | - | 3C-like proteinase | 34.7 | - | No | - | 63.7 | 16.3 | 65.4 | - | |||||||
| 7s70 | 2.60 Å | 2022-07-27 | X-ray | 8H3 | SARS-CoV-2 | NSP5 (3CLpro) | 24.89% | 32.24% | N/A | (1R,2S,5S)-N-{(2S,3R)-4-(butylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(2S,3R)-4-(butylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | iMOSFLM | 99.90% | 4.6 | ALS (5.0.2) | PHENIX | 2021-09-15 | 35839690 | - | - | - | - | N/A | - | - | 3C-like proteinase | 0.8 | - | No | - | 4.8 | 14.3 | 24.0 | - | |||||||
| 7s71 | 1.85 Å | 2022-07-27 | X-ray | 8H9 | SARS-CoV-2 | NSP5 (3CLpro) | 19.45% | 21.86% | N/A | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-(hexylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-(hexylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | PROTEUM PLUS | 100.00% | 16.3 | ROTATING ANODE () | PHENIX | 2021-09-15 | 35839690 | - | - | - | - | N/A | - | - | 3C-like proteinase | 32.2 | - | No | - | 66.5 | 16.3 | 57.6 | - | |||||||
| 7s72 | 2.50 Å | 2022-07-27 | X-ray | 8I0 | SARS-CoV-2 | NSP5 (3CLpro) | 24.11% | 29.58% | N/A | (1R,2S,5S)-N-{(2S,3R)-4-(benzylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(2S,3R)-4-(benzylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | iMOSFLM | 99.40% | 2.7 | ALS (5.0.2) | PHENIX | 2021-09-15 | 35839690 | - | - | - | - | N/A | - | - | 3C-like proteinase | 1.3 | - | No | - | 8.3 | 16.9 | 22.6 | - | |||||||
| 7s73 | 1.85 Å | 2022-07-27 | X-ray | I69 | SARS-CoV-2 | NSP5 (3CLpro) | 20.58% | 23.81% | N/A | (6S)-5-{(2S)-2-[(tert-butylcarbamoyl)amino]-3,3-dimethylbutanoyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-5-azaspiro[2.4]heptane-6-carboxamide (non-preferred name) | (6S)-5-{(2S)-2-[(tert-butylcarbamoyl)amino]-3,3-dimethylbutanoyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-5-azaspiro[2.4]heptane-6-carboxamide (non-preferred name) | PROTEUM PLUS | 99.60% | 18.4 | ROTATING ANODE () | PHENIX | 2021-09-15 | 35839690 | - | - | - | - | N/A | - | - | 3C-like proteinase | 14.1 | - | No | - | 46.7 | 15.6 | 38.2 | - | |||||||
| 7s74 | 1.70 Å | 2022-07-27 | X-ray | I68 | SARS-CoV-2 | NSP5 (3CLpro) | 25.07% | 27.22% | N/A | N-(tert-butylcarbamoyl)-3-methyl-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | N-(tert-butylcarbamoyl)-3-methyl-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | iMOSFLM | 96.30% | 3.5 | ALS (5.0.2) | PHENIX | 2021-09-15 | 35839690 | - | - | - | - | N/A | - | - | 3C-like proteinase | 8.2 | - | No | - | 18.3 | 30.7 | 33.4 | - | |||||||
| 7s75 | 1.80 Å | 2022-07-27 | X-ray | 8I7 | SARS-CoV-2 | NSP5 (3CLpro) | 26.96% | 30.86% | N/A | (1R,2S,5S)-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(3-methylbutanoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(3-methylbutanoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | PROTEUM PLUS | 99.00% | 12.0 | ROTATING ANODE () | PHENIX | 2021-09-15 | 35839690 | - | - | - | - | N/A | - | - | 3C-like proteinase | 5.4 | - | No | - | 6.1 | 13.5 | 52.0 | - | |||||||
| 7wz1 | 3.40 Å | 2022-07-27 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-16 | 35924918 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7fh0 | 3.20 Å | 2022-08-03 | X-ray | NAG | SARS-CoV-2 | Spike | 20.75% | 26.28% | 15.30% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 100.00% | 20.3 | SSRF (BL17U1) | REFMAC | 2021-07-28 | 35906408 | - | - | - | - | N/A | - | - | nanobodies aRBD-2-7; Spike protein S1,Spike protein S1 | 25.9 | - | No | - | 24.7 | 37.0 | 65.7 | - | |||||||
| 7p8i | 4.50 Å | 2022-08-03 | X-ray | NAG | Bat-CoV-RaTG13 | Spike | 27.77% | 29.44% | 27.80% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 99.40% | 3.9 | ESRF (ID23-1) | PHENIX | 2021-07-22 | - | - | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike glycoprotein | - | - | No | - | - | 8.7 | - | 46.7 | - | ||||||
| 7p8j | 6.58 Å | 2022-08-03 | X-ray | - | Bat-CoV-RaTG13 | Spike | 31.79% | 34.50% | 45.90% | - | xia2 | 99.70% | 2.8 | ESRF (ID23-1) | PHENIX | 2021-07-22 | - | - | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike glycoprotein | - | - | No | - | - | 3.9 | - | 42.5 | - | |||||||
| 7qti | 3.04 Å | 2022-08-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-14 | 35879526 | - | - | - | - | N/A | - | - | P5C3 Heavy Chain; P2G3 Light Chain; Spike glycoprotein,Envelope glycoprotein; P5C3 Light Chain; P2G3 Heavy Chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7qtj | 4.01 Å | 2022-08-03 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-01-14 | 35879526 | - | - | - | - | N/A | - | - | P2G3 Heavy Chain; Surface glycoprotein; P5C3 Heavy Chain; P5C3 Light Chain; P2G3 Light Chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7qtk | 3.84 Å | 2022-08-03 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-01-14 | 35879526 | - | - | - | - | N/A | - | - | P2G3 Light Chain; Surface glycoprotein; P2G3 Heavy Chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7t7b | 2.59 Å | 2022-08-03 | X-ray | - | SARS-CoV-2 | Spike | 22.09% | 26.68% | 23.70% | - | HKL-3000 | 99.20% | 5.7 | SSRL (BL12-1) | PHENIX | 2021-12-14 | 35906394 | - | - | - | - | N/A | - | - | ADI-62113 Fab heavy chain; ADI-62113 Fab light chain; Spike protein S1 - Receptor binding domain, UNP residues 333-530 | 34.3 | - | No | - | 21.8 | 63.0 | 59.8 | - | ||||||||
| 7tuu | 1.85 Å | 2022-08-03 | X-ray | U88 | SARS-CoV-2 | NSP5 (3CLpro) | 24.93% | 27.19% | N/A | 5-nitro-1,3-thiazole | 5-nitro-1,3-thiazole | PROTEUM PLUS | 99.40% | 13.0 | ROTATING ANODE () | PHENIX | 2022-02-03 | 35815070 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 10.7 | - | No | - | 18.5 | 13.2 | 58.8 | - | |||||||
| 7vgr | 2.70 Å | 2022-08-03 | Cryo-EM | - | SARS-CoV-2 | Membrane_glycoprotein | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-09-18 | 35931673 | - | - | - | - | N/A | - | - | YN7756_1 Fab heavy chain; Membrane protein; YN7756_1 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7vgs | 2.80 Å | 2022-08-03 | Cryo-EM | - | SARS-CoV-2 | Membrane_glycoprotein | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-09-18 | 35931673 | - | - | - | - | N/A | - | - | YN7717_9 Fab light chain; Membrane protein; YN7717_9 Fab heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7w6m | 4.70 Å | 2022-08-03 | Cryo-EM | NAG | PEDV | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-12-02 | 35986008 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7w73 | 6.40 Å | 2022-08-03 | Cryo-EM | NAG | PEDV | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2021-12-03 | 35986008 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7y6s | 3.10 Å | 2022-08-03 | Cryo-EM | NAG | PEDV | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-06-21 | 35986008 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7y6t | 4.20 Å | 2022-08-03 | Cryo-EM | NAG | PEDV | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-06-21 | 35986008 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7y6u | 3.20 Å | 2022-08-03 | Cryo-EM | NAG | PEDV | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-06-21 | 35986008 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7y6v | 3.30 Å | 2022-08-03 | Cryo-EM | NAG | PEDV | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-06-21 | 35986008 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8df5 | 2.70 Å | 2022-08-03 | X-ray | NAG | SARS-CoV-2 | Spike | 17.74% | 21.71% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 90.80% | 12.3 | SSRL (BL9-2) | PHENIX | 2022-06-21 | 35762884 | ZN | - | - | - | N/A | - | - | S309 Fab Light Chain; S309 Fab Heavy Chain; Spike protein S1; S304 Fab Light Chain; S304 Fab Heavy Chain; Angiotensin-converting enzyme 2 | 71.7 | - | No | - | 67.8 | 82.1 | 69.7 | - | |||||||
| 8dt3 | 3.30 Å | 2022-08-03 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-07-25 | 35926067 | - | - | - | - | N/A | - | - | Light chain Fab of SW186; Spike glycoprotein; Heavy chain Fab of SW186 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7fa1 | 1.60 Å | 2022-08-10 | X-ray | - | SARS-CoV | NSP2 | 18.79% | 22.15% | 7.30% | - | HKL-2000 | 98.70% | 12.0 | SSRF, Photon Factory (BL17U, BL-17A) | PHENIX | 2021-07-05 | - | ZN | - | - | - | N/A | - | - | Non-structural protein 2 - nsp2N | 66.0 | - | No | - | - | 63.7 | 52.4 | 90.7 | - | |||||||
| 7fac | 2.71 Å | 2022-08-10 | X-ray | - | SARS-CoV | NSP2 | 19.72% | 25.70% | 13.10% | - | HKL-2000 | 99.50% | 8.7 | Photon Factory (BL-17A) | PHENIX | 2021-07-06 | - | ZN | - | - | - | N/A | - | - | Non-structural protein 2 - nsp2C | 3.9 | - | No | - | - | 29.0 | 19.6 | 16.0 | - | |||||||
| 7fjc | 2.96 Å | 2022-08-10 | X-ray | - | SARS-CoV-2 | Spike | 21.16% | 26.91% | N/A | - | HKL-2000 | 94.71% | 11.1 | SSRF (BL18U1) | PHENIX | 2021-08-03 | - | - | - | - | - | N/A | - | - | P36-5D2 light chain; P36-5D2 heavy chain; Spike protein S1 | 12.6 | - | No | - | - | 20.1 | 64.1 | 12.1 | - | |||||||
| 7str | 1.50 Å | 2022-08-10 | X-ray | - | SARS-CoV-2 | Nucleocapsid | 16.14% | 20.18% | 7.60% | - | HKL-3000 | 97.40% | 24.6 | APS (19-ID) | PHENIX | 2021-11-15 | - | - | - | - | - | N/A | - | - | Fab S24-1063, Heavy chain; Nucleoprotein - N-terminal RNA binding domain, residues 47-173; Fab S24-1063, Light chain | 65.3 | - | No | - | - | 80.5 | 34.7 | 90.2 | - | |||||||
| 7sts | 2.16 Å | 2022-08-10 | X-ray | - | SARS-CoV-2 | Nucleocapsid | 18.98% | 20.86% | 8.80% | - | HKL-3000; xia2 | 100.00% | 17.5 | APS (19-ID) | PHENIX | 2021-11-15 | - | - | - | - | - | N/A | - | - | Fab S24-1379, heavy chain; Fab S24-1379, light chain; Nucleoprotein - N-terminal RNA binding domain, residues 47-173 | 56.1 | - | No | - | - | 75.2 | 55.7 | 56.0 | - | |||||||
| 7sue | 2.90 Å | 2022-08-10 | X-ray | - | SARS-CoV-2 | Nucleocapsid | 26.17% | 29.19% | 8.50% | - | xia2 | 91.60% | 8.6 | APS (19-ID) | PHENIX | 2021-11-17 | - | - | - | - | - | N/A | - | - | Nucleoprotein - N-terminal RNA binding domain, residues 47-173; S24-188 Fab Light chain; S24-188 Fab Heavy chain | 15.1 | - | No | - | - | 9.4 | 40.0 | 53.5 | - | |||||||
| 7u0p | 3.76 Å | 2022-08-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-18 | 36197988 | - | - | - | - | N/A | - | - | mAb 002_S21F2 light chain; mAb 002-S21F2 Heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7upl | 4.10 Å | 2022-08-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-04-15 | 36197988 | - | - | - | - | N/A | - | - | Spike glycoprotein; mAb 002-S21F2 heavy chain; mAB 002-S21F2 light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7upw | 2.70 Å | 2022-08-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | 42.23% | 42.48% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-04-18 | 35951767 | - | - | - | - | N/A | - | - | Spike glycoprotein; SP1-77 Fab light chain; SP1-77 Fab heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7upx | 3.20 Å | 2022-08-10 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-04-18 | 35951767 | - | - | - | - | N/A | - | - | Spike glycoprotein; SP1-77 Fab light chain; SP1-77 Fab heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7upy | 3.10 Å | 2022-08-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | 42.23% | 42.48% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-04-18 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; SP1-77 Fab heavy chain; SP1-77 Fab light chain | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7v1t | 2.56 Å | 2022-08-10 | X-ray | 5IL | SARS-CoV-2 | NSP5 (3CLpro) | 18.60% | 24.50% | 9.10% | 5,8-bis(oxidanylidene)-7-[(2-piperazin-1-ylphenyl)amino]naphthalene-1-sulfonamide | 5,8-bis(oxidanylidene)-7-[(2-piperazin-1-ylphenyl)amino]naphthalene-1-sulfonamide | HKL-2000 | 99.20% | 14.3 | SSRF (BL19U1) | PHENIX | 2021-08-06 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 47.2 | - | No | - | - | 40.0 | 78.7 | 51.0 | - | ||||||
| 7v20 | 2.86 Å | 2022-08-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-08-07 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7v22 | 3.38 Å | 2022-08-10 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-08-07 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7v23 | 2.96 Å | 2022-08-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-08-07 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; the heavy chain of N12-9 Fab; the light chain of N12-9 Fab | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7v24 | 3.29 Å | 2022-08-10 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-08-07 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; the heavy chain of N12-9 Fab; the lignt chain of N12-9 Fab | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7v27 | 4.18 Å | 2022-08-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-08-07 | - | - | - | - | - | N/A | - | - | Spike protein S1; XG005-VL; XG005-VH | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7voa | 1.80 Å | 2022-08-10 | X-ray | NAG | SARS-CoV-2 | Spike | 18.63% | 22.62% | 10.20% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 97.80% | 13.34 | APS (21-ID-D) | PHENIX | 2021-10-13 | 35906408 | - | - | - | - | N/A | - | - | alpaca nanobody; Spike glycoprotein - UNP residues 321-528 | 33.3 | - | No | - | 58.9 | 23.5 | 60.2 | - | |||||||
| 7wcd | 3.30 Å | 2022-08-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-12-19 | 35931732 | - | - | - | - | N/A | - | - | Spike glycoprotein; Heavy chain; Light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7v3l | 3.47 Å | 2022-08-17 | Cryo-EM | - | MERS-CoV | Spike | N/A | N/A | N/A | - | - | - | () | 2021-08-10 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; antibody L; antibody H | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||||
| 7w8s | 2.85 Å | 2022-08-17 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-12-08 | 36000849 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike protein S1 - Y453F | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7wa1 | 2.90 Å | 2022-08-17 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-12-11 | 36000849 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike protein S1 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7wm0 | 3.35 Å | 2022-08-17 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-01-14 | - | - | - | - | - | N/A | - | - | Heavy chain of 35B5 Fab; Light chain of 35B5 Fab; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7x7t | 3.48 Å | 2022-08-17 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-03-10 | 35972965 | - | - | - | - | N/A | - | - | X01 light chain; X01 heavy chain; X17 heavy chain; X17 light chain; X10 heavy chain; X10 light chain; Spike protein S1 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7x7u | 3.77 Å | 2022-08-17 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-03-10 | 35972965 | - | - | - | - | N/A | - | - | X10 light chain; Spike protein S1; X01 heavy chain; X01 light chain; X17 heavy chain; X17 light chain; X10 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7x7v | 3.83 Å | 2022-08-17 | Cryo-EM | - | SARS-CoV | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | - | () | 2022-03-10 | 35972965 | - | - | - | - | N/A | - | - | X01 heavy chain; Spike protein S1; X10 light chain; X10 heavy chain; X17 light chain; X17 heavy chain; X01 light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7y96 | 3.42 Å | 2022-08-17 | X-ray | - | SARS-CoV-2 | Membrane_glycoprotein | 23.97% | 26.46% | 15.00% | - | XDS | 93.10% | 8.9 | SSRF (BL18U1) | PHENIX | 2022-06-24 | 36874273 | - | - | - | - | N/A | - | - | Green fluorescent protein,Membrane protein - carboxy-terminal domain | 24.6 | - | No | - | 23.3 | 67.1 | 34.3 | - | ||||||||
| 7y9b | 3.21 Å | 2022-08-17 | X-ray | N8E | SARS-CoV-2 | Membrane_glycoprotein | 27.09% | 29.31% | 14.70% | 3,6,9,12,15-PENTAOXATRICOSAN-1-OL | 3,6,9,12,15-PENTAOXATRICOSAN-1-OL | XDS | 94.10% | 6.9 | SSRF (BL18U1) | PHENIX | 2022-06-24 | 36874273 | - | - | - | - | N/A | - | - | Membrane protein | 34.4 | - | No | - | 9.0 | 78.2 | 57.6 | - | |||||||
| 7y9n | 1.89 Å | 2022-08-17 | X-ray | - | SARS-CoV-2 | Spike | 19.99% | 21.24% | 10.20% | SARS-coV-2 S2 subunit | - | HKL-2000 | 99.60% | 26.444 | SSRF (BL19U1) | PHENIX | 2022-06-25 | 35757908 | - | - | - | - | N/A | - | - | Spike protein S2',5HB-H2; SARS-coV-2 S2 subunit - HR2 domain | 42.4 | - | No | - | 72.0 | 19.9 | 68.6 | - | |||||||
| 7zbu | 4.31 Å | 2022-08-17 | Cryo-EM | 3Q9, NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 3-[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-2-[[5-[(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid; 2-acetamido-2-deoxy-beta-D-glucopyranose | 3-[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-2-[[5-[(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid; 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-03-24 | 35981534 | - | - | - | - | N/A | - | - | P008_60 antibody, Heavy chain; P008_60 antibody, Light chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7pfl | 1.80 Å | 2022-08-24 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 17.22% | 21.92% | 9.70% | SODIUM ION | SODIUM ION | XDS | 97.30% | 7.2 | MAX IV (BioMAX) | PHENIX | 2021-08-11 | - | - | - | - | - | N/A | - | - | Replicase polyprotein 1ab | 71.5 | - | No | - | - | 65.8 | 64.3 | 89.1 | - | ||||||
| 7pfm | 2.00 Å | 2022-08-24 | X-ray | 7IL | SARS-CoV-2 | NSP5 (3CLpro) | 18.08% | 22.00% | 9.50% | N-[(1R)-2-(tert-butylamino)-2-oxidanylidene-1-pyridin-3-yl-ethyl]-N-(4-tert-butylphenyl)-1H-imidazole-5-carboxamide | N-[(1R)-2-(tert-butylamino)-2-oxidanylidene-1-pyridin-3-yl-ethyl]-N-(4-tert-butylphenyl)-1H-imidazole-5-carboxamide | XDS | 99.90% | 9.7 | MAX IV (BioMAX) | PHENIX | 2021-08-11 | - | - | - | - | - | N/A | - | - | Replicase polyprotein 1ab | 74.1 | - | No | - | - | 65.0 | 76.2 | 84.5 | - | ||||||
| 7ru1 | 2.80 Å | 2022-08-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-08-16 | 35971553 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ru2 | 3.00 Å | 2022-08-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-08-16 | 35971553 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ru3 | 3.30 Å | 2022-08-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-08-16 | 35971553 | - | - | - | - | N/A | - | - | Spike glycoprotein; CC6.33 IgG heavy chain Fv; CC6.33 IgG Kappa chain Fv | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ru4 | 3.30 Å | 2022-08-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-08-16 | 35971553 | - | - | - | - | N/A | - | - | CC6.33 IgG kappa chain Fv; Spike glycoprotein; CC6.33 IgG heavy chain Fv | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ru5 | 3.60 Å | 2022-08-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-08-16 | 35971553 | - | - | - | - | N/A | - | - | Spike glycoprotein; CC6.30 Fab heavy chain Fv; CC6.30 Fab kappa chain Fv | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ru8 | 3.80 Å | 2022-08-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-08-16 | 35971553 | - | - | - | - | N/A | - | - | CC6.30 Fab Kappa chain Fv; CC6.30 Fab heavy chain Fv; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7t3m | 3.00 Å | 2022-08-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-12-08 | 36192374 | - | - | - | - | N/A | - | - | Antibody 2-7 scFv, variable heavy chain VH; Spike glycoprotein; Antibody 2-7 scFv, variable light chain VL | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7t67 | 3.00 Å | 2022-08-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-12-13 | 36192374 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7us6 | 3.80 Å | 2022-08-24 | Cryo-EM | NAG | CCoV-HuPn-2018 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-04-23 | 35700730 | - | - | - | - | N/A | - | - | Spike glycoprotein - CCoV-HuPn-2018_ Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7usa | 2.80 Å | 2022-08-24 | Cryo-EM | NAG | CCoV-HuPn-2018 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-04-23 | 35700730 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7v5k | 2.80 Å | 2022-08-24 | Cryo-EM | - | MERS-CoV | Spike | N/A | N/A | N/A | - | - | - | () | 2021-08-17 | - | - | - | - | - | N/A | - | - | 0722 L; Spike glycoprotein; 0722 H | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||||
| 7vvp | 1.97 Å | 2022-08-24 | X-ray | 80I | SARS-CoV-2 | NSP5 (3CLpro) | 20.63% | 24.93% | 7.00% | [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | XDS | 93.90% | 20.6 | SSRF (BL02U1) | PHENIX | 2021-11-07 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 30.5 | - | No | - | - | 42.8 | 38.8 | 55.3 | - | ||||||
| 7wqh | 2.32 Å | 2022-08-24 | X-ray | 80I | HCoV-NL63 | NSP5 (3CLpro) | 20.39% | 26.46% | 5.40% | [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | XDS | 93.40% | 7.9 | SSRF (BL10U2) | PHENIX | 2022-01-25 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 47.1 | - | No | - | - | 23.3 | 80.8 | 65.5 | - | ||||||
| 7wqj | 2.75 Å | 2022-08-24 | X-ray | 80I | MERS-CoV | NSP5 (3CLpro) | 22.43% | 28.98% | 7.40% | [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | XDS | 98.50% | 9.2 | SSRF (BL02U1) | PHENIX | 2022-01-25 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 16.4 | - | No | - | - | 10.1 | 68.4 | 27.6 | - | ||||||
| 7ydi | 3.98 Å | 2022-08-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-07-04 | 36151403 | ZN | - | - | - | N/A | - | - | Light chain of R1-32; Processed angiotensin-converting enzyme 2; Spike protein S1 - Spike protomer RBD domain; Heavy chain of R1-32 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7ydy | 4.75 Å | 2022-08-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-07-04 | 36151403 | - | - | - | - | N/A | - | - | Light chain of R1-32 Fab; Heavy chain of R1-32 Fab; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7ye5 | 6.75 Å | 2022-08-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-07-05 | 36151403 | - | - | - | - | N/A | - | - | Spike glycoprotein; Light chain of R1-32 Fab; Heavy chain of R1-32 Fab | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7ye9 | 4.17 Å | 2022-08-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-07-05 | 36151403 | - | - | - | - | N/A | - | - | Spike glycoprotein; Heavy chain of R1-32 Fab; Light chain of R1-32 Fab | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7yeg | 3.73 Å | 2022-08-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-07-05 | 36151403 | ZN | - | - | - | N/A | - | - | heavy chain of R1-32 Fab; light chain of R1-32 Fab; Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8dad | 3.85 Å | 2022-08-24 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-06-13 | - | - | - | - | - | N/A | - | - | AZ090 Fab Heavy Chain; AZ090 Fab Light Chain; Spike glycoprotein - Spike 6P | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 8di5 | 3.04 Å | 2022-08-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-06-28 | 35875685 | - | - | - | - | N/A | - | - | VH F6; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8doy | 1.59 Å | 2022-08-24 | X-ray | PG0, MES, 1PE, T1X | SARS-CoV-2 | NSP5 (3CLpro) | 19.73% | 24.57% | N/A | 2-(2-METHOXYETHOXY)ETHANOL; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID; PENTAETHYLENE GLYCOL; 7-fluoro-N-[(2S)-1-({(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | DIALS | 91.04% | 5.59 | SPring-8 (BL41XU) | REFMAC | 2022-07-14 | 36338434 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 - UNP residues 3264-3564 | 38.2 | - | No | - | 39.6 | 50.1 | 62.7 | - | ||||||||
| 8dpr | 2.00 Å | 2022-08-24 | X-ray | T43 | SARS-CoV-2 | NSP5 (3CLpro) | 18.97% | 24.72% | N/A | 2,2,2-trifluoro-N-{(2S)-1-[(1R,2S,5S)-2-({(2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamothioyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexan-3-yl]-3,3-dimethyl-1-oxobutan-2-yl}acetamide | xia2 | 99.90% | 9.16 | SPring-8 (BL41XU) | REFMAC | 2022-07-16 | 36841831 | ZN | - | - | - | N/A | - | - | 3C-like proteinase nsp5 - UNP residues 3264-3565 | 50.3 | - | No | - | 37.9 | 83.5 | 54.2 | - | ||||||||
| 8dz9 | 1.66 Å | 2022-08-24 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | 20.77% | 25.06% | N/A | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | XDS | 46.10% | 3.7 | LNLS SIRUS (MANACA) | REFMAC | 2022-08-06 | 36775130 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 36.9 | - | No | - | 34.7 | 89.9 | 25.4 | - | |||||||
| 8e1y | 2.48 Å | 2022-08-24 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | 21.66% | 27.23% | N/A | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | XDS | 59.50% | 3.9 | LNLS SIRUS (MANACA) | REFMAC | 2022-08-11 | 36775130 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 26.2 | - | No | - | 18.2 | 61.8 | 47.8 | - | |||||||
| 7vbd | 1.94 Å | 2022-08-31 | X-ray | - | SARS-CoV-2 | Nucleocapsid | 22.26% | 26.23% | 33.00% | - | XDS | 99.30% | 16.1 | SSRF (BL17U1) | PHENIX | 2021-08-31 | - | - | - | - | - | N/A | - | - | Nucleoprotein - N-terminal domain | 24.0 | - | No | - | - | 24.9 | 19.6 | 78.8 | - | |||||||
| 7wbz | 2.42 Å | 2022-08-31 | X-ray | NAG | SARS-CoV-2 | Spike | 18.21% | 22.01% | 19.00% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.90% | 2.6 | SSRF (BL18U1) | PHENIX | 2021-12-17 | 35931732 | - | - | - | - | N/A | - | - | 2303 heavy chain; Spike protein S1 - receptor binding domain; 2303 light chain | 67.9 | - | No | - | 64.9 | 60.8 | 85.4 | - | |||||||
| 7xch | 3.40 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-03-24 | - | ZN | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7xci | 3.20 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-03-24 | - | ZN | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike glycoprotein - UNP residues 333-527 | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7xck | 2.50 Å | 2022-08-31 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-03-24 | - | - | - | - | - | N/A | - | - | Spike protein S1 - UNP residues 322-590; S309 heavy chain; S309 light chain | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7xcp | 3.05 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-03-24 | - | ZN | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Heavy chain of 304 Fab; Lignt chain of 304 Fab; Spike protein S1 - UNP residues 333-527 | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7y9s | 3.00 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-06-26 | 36002453 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7ya1 | 3.11 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-06-27 | 36002453 | ZN | - | - | - | N/A | - | - | Spike protein S1; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7yad | 2.66 Å | 2022-08-31 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2022-06-27 | 36002453 | - | - | - | - | N/A | - | - | S309 neutralizing antibody heavy chain; S309 neutralizing antibody light chain; Spike protein S1 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 8ddi | 2.80 Å | 2022-08-31 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 17.80% | 24.68% | 11.60% | - | HKL-2000 | 98.20% | 12.18 | APS (22-BM) | REFMAC | 2022-06-18 | 37651467 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 60.4 | - | No | - | 38.5 | 93.3 | 63.5 | - | ||||||||
| 8ddm | 2.78 Å | 2022-08-31 | X-ray | K36 | SARS-CoV-2 | NSP5 (3CLpro) | 19.07% | 24.55% | 9.80% | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | HKL-2000 | 97.70% | 13.22 | APS (22-BM) | REFMAC | 2022-06-18 | 37651467 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 40.1 | - | No | - | 39.7 | 68.7 | 47.7 | - | |||||||
| 8dli | 2.56 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-07-08 | 35982054 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dlj | 2.91 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-07-08 | 35982054 | - | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dlk | 3.04 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-07-08 | 35982054 | - | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dll | 2.56 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-07-08 | 35982054 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dlm | 2.89 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-07-08 | 35982054 | - | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dln | 3.04 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-07-08 | 35982054 | - | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dlo | 2.25 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-07-08 | 35982054 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dlp | 2.64 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-07-08 | 35982054 | - | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dlq | 2.77 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-07-08 | 35982054 | - | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dlr | 2.51 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-07-08 | 35982054 | - | - | - | - | N/A | - | - | Spike glycoprotein; Fab 4-8 heavy chain; Fab 4-8 light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dls | 2.66 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-07-08 | 35982054 | - | - | - | - | N/A | - | - | Fab 4A8 light chain; Fab 4A8 heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dlt | 2.40 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-07-08 | 35982054 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dlu | 3.14 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-07-08 | 35982054 | - | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dlv | 3.11 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-07-08 | 35982054 | - | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dlw | 2.16 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-07-08 | 35982054 | - | - | - | - | N/A | - | - | Fab S2M11 light chain; Fab S2M11 heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dlx | 2.45 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-07-08 | 35982054 | - | - | - | - | N/A | - | - | Spike glycoprotein; VH ab6 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dly | 3.00 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-07-08 | 35982054 | - | - | - | - | N/A | - | - | VH ab6; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dlz | 2.57 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-07-08 | 35982054 | - | - | - | - | N/A | - | - | VH ab6; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dm0 | 3.21 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-07-08 | 35982054 | - | - | - | - | N/A | - | - | Spike glycoprotein; VH ab6 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dv1 | 3.40 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-07-27 | 36805129 | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2,Immunoglobulin gamma-1 heavy chain fusion,Immunoglobulin gamma-1 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8dv2 | 3.50 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-07-27 | 36805129 | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2,Immunoglobulin gamma-1 heavy chain fusion | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8dzh | 3.20 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-08-07 | 37518189 | - | - | - | - | N/A | - | - | Spike glycoprotein; antibody MB.02 light chain - Fab variable domain; antibody MB.02 heavy chain - Fab variable domain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dzi | 3.50 Å | 2022-08-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-08-07 | 37518189 | - | - | - | - | N/A | - | - | Spike glycoprotein; antibody MB.02 light chain - Fab variable domain; antibody MB.02 heavy chain - Fab variable domain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7phz | 1.66 Å | 2022-09-07 | X-ray | X77, NA | SARS-CoV-2 | NSP5 (3CLpro) | 16.12% | 18.15% | 8.30% | N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide; SODIUM ION | N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide; SODIUM ION | XDS | 100.00% | 15.8 | ELETTRA (11.2C) | PHENIX; PHENIX | 2021-08-19 | - | - | - | - | - | N/A | - | - | Replicase polyprotein 1ab | 79.3 | - | No | - | - | 91.6 | 64.4 | 84.4 | - | ||||||
| 7tw7 | 1.62 Å | 2022-09-07 | X-ray | SAM | SARS-CoV-2 | NSP14 | 17.89% | 21.79% | 7.00% | S-ADENOSYLMETHIONINE | S-ADENOSYLMETHIONINE | DIALS | 90.00% | 12.4 | NSLS-II (17-ID-2) | PHENIX | 2022-02-06 | 36075969 | ZN | - | - | - | N/A | - | - | Transcription factor ETV6,Proofreading exoribonuclease nsp14 chimera | 71.6 | - | No | - | 67.2 | 87.2 | 65.0 | - | |||||||
| 7tw8 | 1.55 Å | 2022-09-07 | X-ray | SAH | SARS-CoV-2 | NSP14 | 17.60% | 20.56% | 6.80% | S-ADENOSYL-L-HOMOCYSTEINE | S-ADENOSYL-L-HOMOCYSTEINE | DIALS | 95.40% | 13.4 | NSLS-II (17-ID-2) | PHENIX | 2022-02-06 | 36075969 | ZN | - | - | - | N/A | - | - | Transcription factor ETV6,Proofreading exoribonuclease nsp14 chimera | 72.3 | - | No | - | 77.7 | 59.6 | 83.8 | - | |||||||
| 7tw9 | 1.41 Å | 2022-09-07 | X-ray | SFG, EOH, MOH | SARS-CoV-2 | NSP14 | 19.22% | 23.19% | 10.80% | METHANOL; SINEFUNGIN; ETHANOL | SINEFUNGIN; ETHANOL; METHANOL | DIALS | 90.00% | 9.2 | NSLS-II (17-ID-1) | PHENIX | 2022-02-06 | 36075969 | ZN | - | - | - | N/A | - | - | Transcription factor ETV6,Proofreading exoribonuclease nsp14 chimera | 51.6 | - | No | - | 53.3 | 75.4 | 49.4 | - | |||||||
| 8czi | 2.22 Å | 2022-09-07 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2022-05-24 | 36122200 | - | - | - | - | N/A | - | - | Scaffolded Spike protein S2' HR1; Spike protein S2' HR2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 8e5x | 1.70 Å | 2022-09-07 | X-ray | PG4, URR, UO9 | SARS-CoV-2 | NSP5 (3CLpro) | 17.90% | 21.50% | 8.40% | (2~{S})-2-[[(2~{S})-4-methyl-2-[[2-methyl-2-[oxidanyl(phenyl)-$l^{3}-sulfanyl]propoxy]carbonylamino]pentanoyl]amino]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propane-1-sulfonic acid | TETRAETHYLENE GLYCOL; N~2~-(ethoxycarbonyl)-N-{(1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}-L-leucinamide; (2~{S})-2-[[(2~{S})-4-methyl-2-[[2-methyl-2-[oxidanyl(phenyl)-$l^{3}-sulfanyl]propoxy]carbonylamino]pentanoyl]amino]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propane-1-sulfonic acid | XDS | 100.00% | 22.1 | SEALED TUBE () | PHENIX | 2022-08-22 | - | - | - | - | - | N/A | - | - | 3C-like proteinase - Full Length | 55.3 | - | No | - | - | 69.7 | 40.7 | 74.9 | - | ||||||
| 8e61 | 1.85 Å | 2022-09-07 | X-ray | VLU, VM0 | SARS-CoV-2 | NSP5 (3CLpro) | 17.60% | 21.70% | 5.40% | (1R,2S)-2-{[N-({2-[(3-chlorophenyl)sulfanyl]-2-methylpropoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1R,2S)-2-{[N-({2-[(3-chlorophenyl)sulfanyl]-2-methylpropoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; (1S,2S)-2-{[N-({2-[(3-chlorophenyl)sulfanyl]-2-methylpropoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | XDS | 98.60% | 12.4 | NSLS-II (19-ID) | PHENIX | 2022-08-22 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 66.8 | - | No | - | - | 67.9 | 61.7 | 79.1 | - | ||||||
| 8e6b | 1.55 Å | 2022-09-07 | X-ray | URR, UO9 | MERS-CoV | NSP5 (3CLpro) | 17.40% | 19.80% | 4.40% | N~2~-(ethoxycarbonyl)-N-{(1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}-L-leucinamide | N~2~-(ethoxycarbonyl)-N-{(1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}-L-leucinamide; (2~{S})-2-[[(2~{S})-4-methyl-2-[[2-methyl-2-[oxidanyl(phenyl)-$l^{3}-sulfanyl]propoxy]carbonylamino]pentanoyl]amino]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propane-1-sulfonic acid | XDS | 95.30% | 13.9 | NSLS-II (19-ID) | PHENIX | 2022-08-22 | - | - | - | - | - | N/A | - | - | Orf1a protein - Full Length | 72.1 | - | No | - | - | 83.0 | 71.3 | 66.3 | - | ||||||
| 8e6c | 2.70 Å | 2022-09-07 | X-ray | UTU | MERS-CoV | NSP5 (3CLpro) | 20.08% | 25.87% | 14.70% | [2-(3-fluorophenyl)sulfanyl-2-methyl-propyl] ~{N}-[(2~{S})-1-[[3-[(3~{S})-2-$l^{3}-oxidanylidenepyrrolidin-3-yl]-1-$l^{1}-oxidanylsulfonyl-1-oxidanyl-propan-2-yl]-$l^{2}-azanyl]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | [2-(3-fluorophenyl)sulfanyl-2-methyl-propyl] ~{N}-[(2~{S})-1-[[3-[(3~{S})-2-$l^{3}-oxidanylidenepyrrolidin-3-yl]-1-$l^{1}-oxidanylsulfonyl-1-oxidanyl-propan-2-yl]-$l^{2}-azanyl]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | XDS | 99.70% | 5.3 | NSLS-II (19-ID) | PHENIX | 2022-08-22 | - | - | - | - | - | N/A | - | - | Orf1a protein | 52.4 | - | No | - | - | 27.7 | 88.6 | 63.4 | - | ||||||
| 8e6d | 2.70 Å | 2022-09-07 | X-ray | UTL | MERS-CoV | NSP5 (3CLpro) | 21.03% | 26.38% | 12.90% | (1R,2S)-2-{[N-({2-[(4-fluorophenyl)sulfanyl]-2-methylpropoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1R,2S)-2-{[N-({2-[(4-fluorophenyl)sulfanyl]-2-methylpropoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | XDS | 96.40% | 6.6 | NSLS-II (19-ID) | PHENIX | 2022-08-22 | - | - | - | - | - | N/A | - | - | Orf1a protein | 32.2 | - | No | - | - | 24.0 | 68.5 | 47.9 | - | ||||||
| 8e6e | 1.50 Å | 2022-09-07 | X-ray | UV2, UUR | MERS-CoV | NSP5 (3CLpro) | 17.30% | 20.30% | 5.00% | 2-phenylsulfanylethyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-[(N-{[2-(phenylsulfanyl)ethoxy]carbonyl}-L-leucyl)amino]propane-1-sulfonic acid; 2-phenylsulfanylethyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | XDS | 99.70% | 11.6 | NSLS-II (19-ID) | PHENIX | 2022-08-22 | - | - | - | - | - | N/A | - | - | Orf1a protein | 73.4 | - | No | - | - | 79.6 | 64.4 | 79.8 | - | ||||||
| 7f60 | 2.85 Å | 2022-09-14 | X-ray | - | SARS-CoV-2 | ORF6 | 23.57% | 27.56% | 37.00% | - | XDS | 99.00% | 3.47 | SSRF (BL17B1) | PHENIX | 2021-06-23 | - | - | - | - | - | N/A | - | - | Nuclear pore complex protein Nup98-Nup96; mRNA export factor; ORF6 protein | 25.7 | - | No | - | - | 16.4 | 60.7 | 49.8 | - | |||||||
| 7f90 | 2.39 Å | 2022-09-14 | X-ray | - | SARS-CoV | ORF6 | 21.96% | 25.81% | 13.10% | - | XDS | 99.70% | 7.62 | SSRF (BL19U1) | PHENIX | 2021-07-03 | - | - | - | - | - | N/A | - | - | Nuclear pore complex protein Nup98-Nup96; ORF6 protein; mRNA export factor | 12.8 | - | No | - | - | 28.1 | 30.7 | 38.0 | - | |||||||
| 7t2t | 1.45 Å | 2022-09-14 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 15.51% | 20.01% | 7.20% | - | XDS | 98.40% | 15.98 | SSRL (BL12-2) | REFMAC | 2021-12-06 | 36075915 | - | - | - | - | N/A | - | - | 3C-like proteinase | 65.5 | - | No | - | 81.6 | 56.0 | 68.2 | - | ||||||||
| 7t2u | 2.10 Å | 2022-09-14 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 24.51% | 29.99% | 9.70% | - | XDS | 96.40% | 8.86 | SSRL (BL12-2) | REFMAC | 2021-12-06 | 36075915 | - | - | - | - | N/A | - | - | 3C-Like Protease; NEMO peptide | 13.3 | - | No | - | 7.3 | 37.2 | 53.8 | - | ||||||||
| 7t2v | 2.47 Å | 2022-09-14 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 19.57% | 23.70% | 18.10% | - | XDS | 96.80% | 8.33 | SSRL (BL12-2) | REFMAC | 2021-12-06 | 36075915 | - | - | - | - | N/A | - | - | 3C-Like Protease | 48.7 | - | No | - | 47.7 | 67.7 | 57.2 | - | ||||||||
| 7v6n | 3.99 Å | 2022-09-14 | Cryo-EM | - | MERS-CoV | Spike | N/A | N/A | N/A | - | - | - | () | 2021-08-20 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; 111 L; 111 H | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||||
| 7xxl | 7.30 Å | 2022-09-14 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-05-30 | 36138032 | - | - | - | - | N/A | - | - | Spike protein S1 - RBD; Fab14 light chain; Fab14 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8e64 | 1.75 Å | 2022-09-14 | X-ray | WEQ, WEL | SARS-CoV-2 | NSP5 (3CLpro) | 17.40% | 21.20% | 6.10% | (1S,2S)-2-{[N-({2-[(1H-benzimidazol-2-yl)sulfanyl]-2-methylpropoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1~{R},2~{S})-2-[[(2~{S})-2-[[2-(1~{H}-benzimidazol-2-ylsulfanyl)-2-methyl-propoxy]carbonylamino]-4-methyl-pentanoyl]amino]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propane-1-sulfonic acid;molecular oxygen; (1S,2S)-2-{[N-({2-[(1H-benzimidazol-2-yl)sulfanyl]-2-methylpropoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | XDS | 99.90% | 22.4 | SEALED TUBE () | PHENIX | 2022-08-22 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 81.1 | - | No | - | - | 72.3 | 91.0 | 83.1 | - | ||||||
| 8e65 | 1.80 Å | 2022-09-14 | X-ray | WF5 | SARS-CoV-2 | NSP5 (3CLpro) | 18.10% | 23.50% | 13.00% | (1S,2S)-2-[(N-{[2-(4-chlorophenoxy)-2-methylpropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-[(N-{[2-(4-chlorophenoxy)-2-methylpropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | XDS | 100.00% | 10.1 | NSLS-II (19-ID) | PHENIX | 2022-08-22 | - | - | - | - | - | N/A | - | - | 3C-like proteinase - Full Length | 75.9 | - | No | - | - | 49.8 | 88.4 | 91.9 | - | ||||||
| 8e68 | 1.60 Å | 2022-09-14 | X-ray | WGU, WGO, PG4 | SARS-CoV-2 | NSP5 (3CLpro) | 18.20% | 21.70% | 7.40% | N~2~-{[2-(4-fluorophenoxy)-2-methylpropoxy]carbonyl}-N-{(1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}-L-leucinamide; (1S,2S)-2-[(N-{[2-(4-fluorophenoxy)-2-methylpropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-[(N-{[2-(4-fluorophenoxy)-2-methylpropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; N~2~-{[2-(4-fluorophenoxy)-2-methylpropoxy]carbonyl}-N-{(1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}-L-leucinamide; TETRAETHYLENE GLYCOL | XDS | 99.80% | 12.0 | APS (17-ID) | PHENIX | 2022-08-22 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 56.9 | - | No | - | - | 67.9 | 39.2 | 81.2 | - | ||||||
| 7v5j | 2.80 Å | 2022-09-21 | Cryo-EM | - | MERS-CoV | Spike | N/A | N/A | N/A | - | - | - | () | 2021-08-17 | - | - | - | - | - | N/A | - | - | 0722 L; 0722 H; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||||
| 7xco | 2.50 Å | 2022-09-21 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-03-24 | - | - | - | - | - | N/A | - | - | S309 fab light chain; Spike glycoprotein; S309 Fab heavy chain | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7y9z | 2.85 Å | 2022-09-21 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-06-26 | 36002453 | ZN | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7ya0 | 3.10 Å | 2022-09-21 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-06-26 | 36002453 | ZN | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dox | 1.46 Å | 2022-09-21 | X-ray | T2L | SARS-CoV-2 | NSP5 (3CLpro) | 19.93% | 22.89% | 17.80% | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | xia2 | 99.10% | 8.68 | SPring-8 (BL24XU) | REFMAC | 2022-07-14 | 36841831 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 - UNP residues 3264-3565 | 63.8 | - | No | - | 56.2 | 68.6 | 77.4 | - | |||||||
| 8drr | 2.00 Å | 2022-09-21 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 20.59% | 23.71% | 8.70% | SODIUM ION | SODIUM ION | XDS | 100.00% | 12.8 | APS (23-ID-D) | PHENIX | 2022-07-21 | 36057636 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 - UNP residues 3264-3563 | 16.9 | - | No | - | 47.7 | 23.1 | 36.5 | - | |||||||
| 8drs | 1.80 Å | 2022-09-21 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 17.97% | 21.79% | 19.00% | - | iMOSFLM | 99.80% | 7.0 | ALS (5.0.2) | PHENIX | 2022-07-21 | 36057636 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 - UNP residues 3264-3563 | 41.2 | - | No | - | 67.2 | 43.3 | 47.7 | - | ||||||||
| 8drt | 1.50 Å | 2022-09-21 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 15.46% | 18.27% | 4.60% | - | autoPROC | 68.00% | 16.8 | APS (23-ID-B) | PHENIX | 2022-07-21 | 36057636 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 - UNP residues 3264-3563 | 77.1 | - | No | - | 91.1 | 58.1 | 84.5 | - | ||||||||
| 8dru | 2.31 Å | 2022-09-21 | X-ray | 1PE, PO4 | SARS-CoV-2 | NSP5 (3CLpro) | 19.00% | 23.71% | 7.30% | PENTAETHYLENE GLYCOL; PHOSPHATE ION | PENTAETHYLENE GLYCOL; PHOSPHATE ION | XDS | 93.10% | 11.2 | CLSI (08B1-1) | PHENIX | 2022-07-21 | 36057636 | - | - | - | - | N/A | - | - | Fusion protein of 3C-like proteinase nsp5 and nsp7-nsp8 (C7) cut site | 13.8 | - | No | - | 47.7 | 30.8 | 21.2 | - | |||||||
| 8drv | 2.40 Å | 2022-09-21 | X-ray | 1PE | SARS-CoV-2 | NSP5 (3CLpro) | 17.76% | 23.98% | 18.10% | PENTAETHYLENE GLYCOL | PENTAETHYLENE GLYCOL | XDS | 99.90% | 9.9 | APS (23-ID-B) | PHENIX | 2022-07-21 | 36057636 | - | - | - | - | N/A | - | - | Fusion protein of 3C-like proteinase nsp5 and nsp8-nsp9 (C8) cut site | 54.1 | - | No | - | 45.2 | 91.1 | 46.6 | - | |||||||
| 8drw | 2.67 Å | 2022-09-21 | X-ray | PO4, NA, 1PE | SARS-CoV-2 | NSP5 (3CLpro) | 20.46% | 23.92% | 17.90% | SODIUM ION; PHOSPHATE ION; PENTAETHYLENE GLYCOL | PHOSPHATE ION; SODIUM ION; PENTAETHYLENE GLYCOL | autoPROC | 99.40% | 7.3 | APS (23-ID-B) | PHENIX | 2022-07-21 | 36057636 | - | - | - | - | N/A | - | - | Fusion protein of 3C-like proteinase nsp5 and nsp9-nsp10 (C9) cut site | 20.6 | - | No | - | 45.6 | 40.4 | 30.0 | - | |||||||
| 8drx | 1.50 Å | 2022-09-21 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 16.26% | 19.16% | 5.20% | SODIUM ION | SODIUM ION | 95.10% | 17.7 | CLSI (08B1-1) | PHENIX | 2022-07-21 | 36057636 | - | - | - | - | N/A | - | - | Fusion protein of 3C-like proteinase nsp5 and nsp10-nsp11 (C10) cut site | 56.4 | - | No | - | 86.9 | 40.0 | 60.5 | - | ||||||||
| 8dry | 2.49 Å | 2022-09-21 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 19.42% | 26.41% | N/A | - | autoPROC | 72.60% | 9.1 | APS (23-ID-B) | PHENIX | 2022-07-21 | 36057636 | - | - | - | - | N/A | - | - | Fusion protein of 3C-like proteinase nsp5 and nsp12-nsp13 (C12) cut site | 22.9 | - | No | - | 23.6 | 76.6 | 20.9 | - | ||||||||
| 8e5z | 1.80 Å | 2022-09-21 | X-ray | UQO, WJB | SARS-CoV-2 | NSP5 (3CLpro) | 19.50% | 25.30% | 6.60% | (1R,2S)-2-[(N-{[2-(benzenesulfonyl)-2-methylpropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-[(N-{[2-(benzenesulfonyl)-2-methylpropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; (1R,2S)-2-[(N-{[2-(benzenesulfonyl)-2-methylpropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | XDS | 98.50% | 11.0 | APS (17-ID) | PHENIX | 2022-08-22 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 64.9 | - | No | - | - | 32.4 | 91.0 | 81.3 | - | ||||||
| 8e69 | 2.26 Å | 2022-09-21 | X-ray | WIO | SARS-CoV-2 | NSP5 (3CLpro) | 19.70% | 26.50% | 6.60% | (1R,2S)-2-[(N-{[2-(3-fluorophenoxy)-2-methylpropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1R,2S)-2-[(N-{[2-(3-fluorophenoxy)-2-methylpropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | XDS | 99.50% | 9.2 | APS (17-ID) | PHENIX | 2022-08-22 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 48.9 | - | No | - | - | 22.9 | 73.4 | 76.8 | - | ||||||
| 8e6a | 2.05 Å | 2022-09-21 | X-ray | WIX, WJ0 | SARS-CoV-2 | NSP5 (3CLpro) | 20.20% | 27.70% | 6.70% | (1S,2S)-2-[(N-{[(2R)-2-(3-chlorophenyl)-2-hydroxypropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-[(N-{[(2R)-2-(3-chlorophenyl)-2-hydroxypropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; (1R,2S)-2-[(N-{[(2S)-2-(3-chlorophenyl)-2-hydroxypropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl]propane-1-sulfonic acid | XDS | 99.40% | 10.3 | APS (17-ID) | PHENIX | 2022-08-22 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 47.0 | - | No | - | - | 15.5 | 90.8 | 62.9 | - | ||||||
| 7v6o | 4.56 Å | 2022-09-28 | Cryo-EM | - | MERS-CoV | Spike | N/A | N/A | N/A | - | - | - | () | 2021-08-20 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; 111 L; 111 H | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||||
| 7wrj | 4.08 Å | 2022-09-28 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-01-26 | - | - | - | - | - | N/A | - | - | Spike protein S1; BD55-4637L; BD55-4637H | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7wry | 3.28 Å | 2022-09-28 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-01-27 | 36493787 | - | - | - | - | N/A | - | - | BD55-3546L; BD55-3546H; Spike protein S1 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7xwa | 3.36 Å | 2022-09-28 | X-ray | NAG | SARS-CoV-2 | Spike | 23.09% | 28.67% | N/A | alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 98.80% | 6.7 | Photon Factory (BL-17A) | BUSTER; PHENIX | 2022-05-26 | 36198317 | ZN | - | - | - | N/A | - | - | Spike protein S1; Processed angiotensin-converting enzyme 2 | 8.1 | - | No | - | 11.2 | 20.9 | 50.0 | - | |||||||
| 7y0c | 2.94 Å | 2022-09-28 | X-ray | - | SARS-CoV-2 | Spike | 20.20% | 27.78% | N/A | - | HKL-2000 | 99.61% | 1.62 | SSRF (BL17U) | PHENIX | 2022-06-04 | 36493787 | - | - | - | - | N/A | - | - | BD55-1403 Fab heavy chain; Spike protein S1 - Omicron RBD; BD55-1403 Fab light chain | 14.4 | - | No | - | 15.3 | 56.1 | 29.7 | - | ||||||||
| 7y0v | 2.48 Å | 2022-09-28 | X-ray | - | SARS-CoV-2 | Spike | 21.09% | 26.76% | 10.40% | - | xia2 | 99.90% | 12.98 | SSRF (BL10U2) | PHENIX | 2022-06-06 | 36493787 | - | - | - | - | N/A | - | - | 5549-Fab; 5549-Fab; Spike protein S1 - BA.1-RBD | 11.0 | - | No | - | 21.2 | 28.7 | 41.6 | - | ||||||||
| 7y0w | 3.42 Å | 2022-09-28 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2022-06-06 | 36493787 | - | - | - | - | N/A | - | - | Spike protein S1; BD55-5514L; BD55-5514H; BD55-5840L; BD55-5840H | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 8acd | 1.39 Å | 2022-09-28 | X-ray | LQ6 | SARS-CoV-2 | NSP5 (3CLpro) | 14.54% | 18.37% | 5.70% | (2~{S})-4-[[2,4-bis(oxidanylidene)-1~{H}-pyrimidin-6-yl]carbonyl]-1-(3,4-dichlorophenyl)-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide | (2~{S})-4-[[2,4-bis(oxidanylidene)-1~{H}-pyrimidin-6-yl]carbonyl]-1-(3,4-dichlorophenyl)-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide | XDS | 82.80% | 14.9 | PETRA III, EMBL c/o DESY (P14 (MX2)) | PHENIX | 2022-07-05 | 36107752 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 71.6 | - | No | - | 90.8 | 51.8 | 76.9 | - | |||||||
| 8acl | 1.40 Å | 2022-09-28 | X-ray | LQL | SARS-CoV-2 | NSP5 (3CLpro) | 13.80% | 17.23% | 6.10% | (2~{S})-1-(3,4-dichlorophenyl)-4-pyridin-3-ylcarbonyl-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide | (2~{S})-1-(3,4-dichlorophenyl)-4-pyridin-3-ylcarbonyl-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide | XDS | 86.50% | 13.1 | PETRA III, EMBL c/o DESY (P13 (MX1)) | PHENIX | 2022-07-05 | 36107752 | - | - | - | - | N/A | - | - | Replicase polyprotein 1ab | 65.7 | - | No | - | 94.5 | 39.4 | 72.4 | - | |||||||
| 8drz | 1.98 Å | 2022-09-28 | X-ray | 1PE, TRS, NA | SARS-CoV-2 | NSP5 (3CLpro) | 17.68% | 22.33% | 13.40% | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL; PENTAETHYLENE GLYCOL; SODIUM ION | PENTAETHYLENE GLYCOL; 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL; SODIUM ION | 100.00% | 10.7 | ALS (5.0.1) | PHENIX | 2022-07-21 | 36057636 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 30.0 | - | No | - | 61.9 | 27.9 | 46.2 | - | ||||||||
| 8ds0 | 2.20 Å | 2022-09-28 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 19.29% | 24.59% | 19.40% | - | 99.90% | 8.7 | APS (23-ID-B) | PHENIX | 2022-07-21 | 36057636 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 13.7 | - | No | - | 39.4 | 37.5 | 22.3 | - | |||||||||
| 8ds1 | 2.19 Å | 2022-09-28 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 19.79% | 24.84% | 13.40% | SODIUM ION | SODIUM ION | 99.80% | 10.9 | CLSI (08B1-1) | PHENIX | 2022-07-21 | 36057636 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 16.6 | - | No | - | 36.7 | 30.1 | 39.8 | - | ||||||||
| 8ds2 | 1.60 Å | 2022-09-28 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 15.40% | 18.68% | 7.60% | SODIUM ION | SODIUM ION | 96.94% | 17.12 | ALS (5.0.2) | PHENIX | 2022-07-21 | 36057636 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 79.5 | - | No | - | 89.4 | 71.3 | 80.3 | - | ||||||||
| 8e63 | 1.75 Å | 2022-09-28 | X-ray | UV2, UUR, PG4 | SARS-CoV-2 | NSP5 (3CLpro) | 17.90% | 21.70% | 6.30% | 2-phenylsulfanylethyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate; (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-[(N-{[2-(phenylsulfanyl)ethoxy]carbonyl}-L-leucyl)amino]propane-1-sulfonic acid | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-[(N-{[2-(phenylsulfanyl)ethoxy]carbonyl}-L-leucyl)amino]propane-1-sulfonic acid; 2-phenylsulfanylethyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate; TETRAETHYLENE GLYCOL | XDS | 99.90% | 9.6 | NSLS-II (19-ID) | PHENIX | 2022-08-22 | - | - | - | - | - | N/A | - | - | 3C-like proteinase - Full Length | 73.5 | - | No | - | - | 67.9 | 86.0 | 70.2 | - | ||||||
| 8ehj | 2.28 Å | 2022-09-28 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 21.44% | 25.01% | 11.20% | - | HKL-2000 | 98.30% | 21.23 | APS (19-ID) | REFMAC | 2022-09-14 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 44.2 | - | No | - | - | 35.0 | 41.2 | 87.7 | - | |||||||
| 8ehk | 2.18 Å | 2022-09-28 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 17.85% | 23.26% | 12.90% | - | HKL-2000 | 98.60% | 18.68 | APS (19-ID) | REFMAC | 2022-09-14 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 70.5 | - | No | - | - | 52.5 | 88.6 | 75.8 | - | |||||||
| 8ehl | 2.19 Å | 2022-09-28 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 20.72% | 25.38% | 10.00% | - | HKL-2000 | 99.30% | 24.79 | APS (19-ID) | REFMAC | 2022-09-14 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 60.3 | - | No | - | - | 31.9 | 68.5 | 94.6 | - | |||||||
| 8ehm | 1.84 Å | 2022-09-28 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 19.47% | 24.29% | 9.00% | - | HKL-2000 | 99.50% | 26.64 | APS (19-ID) | REFMAC | 2022-09-14 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 59.7 | - | No | - | - | 42.2 | 57.7 | 94.0 | - | |||||||
| 7set | 1.70 Å | 2022-10-05 | X-ray | I70 | SARS-CoV-2 | NSP5 (3CLpro) | 16.70% | 20.56% | N/A | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | XDS | 95.00% | 9.2 | SSRL (BL12-2) | REFMAC | 2021-10-01 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 72.3 | - | No | - | - | 77.7 | 73.8 | 69.6 | - | ||||||
| 7sf1 | 1.85 Å | 2022-10-05 | X-ray | 8ZI | SARS-CoV-2 | NSP5 (3CLpro) | 16.30% | 20.79% | N/A | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(3,3-dimethylbutanoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(3,3-dimethylbutanoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | XDS | 98.80% | 6.9 | SSRL (BL12-1) | REFMAC | 2021-10-02 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 75.0 | - | No | - | - | 75.9 | 83.7 | 68.3 | - | ||||||
| 7sf3 | 1.75 Å | 2022-10-05 | X-ray | 90H | SARS-CoV-2 | NSP5 (3CLpro) | 16.90% | 19.49% | N/A | (1R,2S,5S)-N-{(2S,3R)-3-hydroxy-4-(methylamino)-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(2S,3R)-3-hydroxy-4-(methylamino)-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | XDS | 98.60% | 9.6 | SSRL (BL12-2) | REFMAC | 2021-10-02 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 80.0 | - | No | - | - | 85.1 | 78.7 | 79.0 | - | ||||||
| 7sfb | 1.90 Å | 2022-10-05 | X-ray | PGE, 90U | SARS-CoV-2 | NSP5 (3CLpro) | 18.34% | 23.15% | N/A | benzyl (1R,2S,5S)-2-({(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}carbamoyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-3-carboxylate; TRIETHYLENE GLYCOL | TRIETHYLENE GLYCOL; benzyl (1R,2S,5S)-2-({(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}carbamoyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-3-carboxylate | XDS | 98.30% | 16.7 | SSRL (BL9-2) | REFMAC | 2021-10-03 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 63.0 | - | No | - | - | 53.6 | 73.4 | 73.8 | - | ||||||
| 7sfh | 1.40 Å | 2022-10-05 | X-ray | 91I, CA | SARS-CoV-2 | NSP5 (3CLpro) | 16.49% | 19.31% | N/A | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-(3-phenylpropanoyl)-3-azabicyclo[3.1.0]hexane-2-carboxamide; CALCIUM ION | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-(3-phenylpropanoyl)-3-azabicyclo[3.1.0]hexane-2-carboxamide; CALCIUM ION | XDS | 99.60% | 12.8 | SSRL (BL9-2) | REFMAC | 2021-10-03 | - | CA | - | - | - | N/A | - | - | 3C-like proteinase | 76.2 | - | No | - | - | 86.1 | 73.8 | 71.1 | - | ||||||
| 7sfi | 1.95 Å | 2022-10-05 | X-ray | CA, 91Z | SARS-CoV-2 | NSP5 (3CLpro) | 19.52% | 23.86% | N/A | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[N-(2,4,6-trifluorophenyl)glycyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide; CALCIUM ION | CALCIUM ION; (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[N-(2,4,6-trifluorophenyl)glycyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | XDS | 98.80% | 13.0 | SSRL (BL9-2) | REFMAC | 2021-10-03 | - | CA | - | - | - | N/A | - | - | 3C-like proteinase | 62.4 | - | No | - | - | 46.3 | 78.7 | 74.4 | - | ||||||
| 7sgh | 1.85 Å | 2022-10-05 | X-ray | 99W | SARS-CoV-2 | NSP5 (3CLpro) | 19.48% | 23.15% | N/A | (S)-N-((S)-1-imino-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)-4-methyl-2-(2-((2,4,6-trifluorophenyl)amino)acetamido)pentanamide | (S)-N-((S)-1-imino-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)-4-methyl-2-(2-((2,4,6-trifluorophenyl)amino)acetamido)pentanamide | DIALS | 96.80% | 5.1 | SSRL (BL12-2) | REFMAC | 2021-10-05 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 56.0 | - | No | - | - | 53.6 | 61.1 | 72.0 | - | ||||||
| 7uj9 | 2.25 Å | 2022-10-05 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 20.43% | 25.02% | 13.70% | - | CrysalisPro | 99.70% | 11.05 | ROTATING ANODE () | PHENIX | 2022-03-30 | 36114420 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 - catalytic domain (MPro1-199) | 18.1 | - | No | - | 34.9 | 20.4 | 54.9 | - | ||||||||
| 7ujg | 1.80 Å | 2022-10-05 | X-ray | K36 | SARS-CoV-2 | NSP5 (3CLpro) | 14.23% | 17.92% | 3.80% | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | CrysalisPro | 95.10% | 32.3 | ROTATING ANODE () | PHENIX | 2022-03-30 | 36114420 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 - catalytic domain (MPro1-196) | 72.2 | - | No | - | 92.4 | 47.7 | 80.7 | - | |||||||
| 7uju | 1.85 Å | 2022-10-05 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | 15.40% | 17.45% | 7.20% | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | CrysalisPro | 99.30% | 15.32 | ROTATING ANODE () | PHENIX | 2022-03-31 | 36114420 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 - catalytic domain (MPro1-196) | 69.5 | - | No | - | 94.0 | 38.7 | 82.0 | - | ||||||||
| 7ukk | 2.00 Å | 2022-10-05 | X-ray | K36 | SARS-CoV-2 | NSP5 (3CLpro) | 16.09% | 18.14% | 7.10% | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | CrysalisPro | 99.90% | 19.24 | ROTATING ANODE () | PHENIX | 2022-04-01 | 36114420 | - | - | - | - | N/A | - | - | 3C-like proteinase | 80.7 | - | No | - | 91.6 | 73.8 | 79.6 | - | |||||||
| 7vib | 3.20 Å | 2022-10-05 | X-ray | - | Pangolin CoV | Spike | 20.36% | 25.79% | 10.70% | - | HKL-2000 | 98.40% | 5.2 | SSRF (BL10U2) | PHENIX | 2021-09-26 | - | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | 23.8 | - | No | - | - | 28.4 | 63.4 | 31.0 | - | |||||||
| 7xrp | 3.88 Å | 2022-10-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-05-11 | 36109732 | - | - | - | - | N/A | - | - | Spike protein S1 - RBD; C5G2 nanobody; Spike protein S1 - NTD | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dz0 | 2.29 Å | 2022-10-05 | X-ray | 7YY | SARS-CoV-2 | NSP5 (3CLpro) | 21.90% | 27.53% | N/A | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | XDS | 89.40% | 4.3 | LNLS SIRUS (MANACA) | PHENIX | 2022-08-06 | 36775130 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 10.0 | - | No | - | 16.5 | 45.4 | 26.4 | - | |||||||
| 8dz1 | 2.08 Å | 2022-10-05 | X-ray | 7YY | SARS-CoV-2 | NSP5 (3CLpro) | 21.79% | 25.61% | N/A | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | XDS | 90.10% | 6.2 | LNLS SIRUS (MANACA) | PHENIX | 2022-08-06 | 36775130 | - | - | - | - | N/A | - | - | Replicase polyprotein 1ab | 15.0 | - | No | - | 29.8 | 36.1 | 36.7 | - | |||||||
| 8dz2 | 2.13 Å | 2022-10-05 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | 18.91% | 22.95% | N/A | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | XDS | 98.60% | 8.6 | LNLS SIRUS (MANACA) | REFMAC | 2022-08-06 | 36775130 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 63.6 | - | No | - | 55.5 | 86.1 | 60.2 | - | |||||||
| 8dz6 | 2.37 Å | 2022-10-05 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | 22.86% | 29.61% | N/A | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | XDS | 56.90% | 4.6 | LNLS SIRUS (MANACA) | REFMAC | 2022-08-06 | 36775130 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 17.9 | - | No | - | 8.2 | 66.4 | 35.0 | - | |||||||
| 8dza | 1.96 Å | 2022-10-05 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | 20.07% | 24.19% | N/A | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | XDS | 58.70% | 7.8 | LNLS SIRUS (MANACA) | REFMAC | 2022-08-06 | 36775130 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 36.6 | - | No | - | 43.2 | 53.7 | 52.4 | - | |||||||
| 7mat | 2.74 Å | 2022-10-12 | X-ray | H37 | SARS-CoV-2 | NSP5 (3CLpro) | 19.55% | 25.95% | 6.10% | D-phenylalanyl-N-[(3S)-6-carbamimidamido-1-chloro-2-oxohexan-3-yl]-L-phenylalaninamide | D-phenylalanyl-N-[(3S)-6-carbamimidamido-1-chloro-2-oxohexan-3-yl]-L-phenylalaninamide | HKL-2000 | 98.20% | 16.9 | ROTATING ANODE () | PHENIX | 2021-03-31 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 25.7 | - | No | - | - | 27.0 | 42.6 | 57.2 | - | ||||||
| 7mau | 1.95 Å | 2022-10-12 | X-ray | DIO, YVP | SARS-CoV-2 | NSP5 (3CLpro) | 17.10% | 21.45% | 5.00% | N-{3-[(prop-2-yn-1-yl)oxy]propanoyl}-D-phenylalanyl-N-[(3S)-6-carbamimidamido-2-oxohexan-3-yl]-L-phenylalaninamide | 1,4-DIETHYLENE DIOXIDE; N-{3-[(prop-2-yn-1-yl)oxy]propanoyl}-D-phenylalanyl-N-[(3S)-6-carbamimidamido-2-oxohexan-3-yl]-L-phenylalaninamide | HKL-2000 | 97.80% | 17.5 | ROTATING ANODE () | PHENIX | 2021-03-31 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 62.5 | - | No | - | - | 70.2 | 50.2 | 79.3 | - | ||||||
| 7mav | 1.91 Å | 2022-10-12 | X-ray | YVY | SARS-CoV-2 | NSP5 (3CLpro) | 20.09% | 24.45% | 11.00% | N-{3-[(prop-2-yn-1-yl)oxy]propanoyl}-D-phenylalanyl-N-[(3S)-6-(carbamoylamino)-2-oxohexan-3-yl]-L-phenylalaninamide | N-{3-[(prop-2-yn-1-yl)oxy]propanoyl}-D-phenylalanyl-N-[(3S)-6-(carbamoylamino)-2-oxohexan-3-yl]-L-phenylalaninamide | XDS | 97.20% | 8.3 | NSLS-II (17-ID-2) | PHENIX | 2021-03-31 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 45.7 | - | No | - | - | 40.6 | 73.6 | 52.5 | - | ||||||
| 7maw | 2.07 Å | 2022-10-12 | X-ray | YVA | SARS-CoV-2 | NSP5 (3CLpro) | 17.21% | 22.86% | 3.60% | ethyl (4R)-4-({3-cyclopropyl-N-[(2E)-3-(4-ethynylphenyl)prop-2-enoyl]-L-alanyl}amino)-5-[(3S)-2-oxopyrrolidin-3-yl]pentanoate | ethyl (4R)-4-({3-cyclopropyl-N-[(2E)-3-(4-ethynylphenyl)prop-2-enoyl]-L-alanyl}amino)-5-[(3S)-2-oxopyrrolidin-3-yl]pentanoate | 98.90% | 12.3 | NSLS-II (17-ID-2) | PHENIX | 2021-03-31 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 55.8 | - | No | - | - | 56.5 | 60.1 | 69.8 | - | |||||||
| 7max | 1.98 Å | 2022-10-12 | X-ray | YV7 | SARS-CoV-2 | NSP5 (3CLpro) | 20.13% | 24.30% | 2.60% | D-phenylalanyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-4-fluoro-L-phenylalaninamide | D-phenylalanyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-4-fluoro-L-phenylalaninamide | 99.40% | 13.7 | NSLS-II (17-ID-2) | PHENIX | 2021-03-31 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 18.4 | - | No | - | - | 41.9 | 15.7 | 53.2 | - | |||||||
| 7maz | 1.70 Å | 2022-10-12 | X-ray | YVD | SARS-CoV-2 | NSP5 (3CLpro) | 16.40% | 20.75% | 4.40% | 4-fluoro-D-phenylalanyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-4-fluoro-L-phenylalaninamide | 4-fluoro-D-phenylalanyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-4-fluoro-L-phenylalaninamide | 95.90% | 10.2 | NSLS-II (17-ID-2) | PHENIX | 2021-03-31 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 65.9 | - | No | - | - | 76.2 | 52.2 | 78.4 | - | |||||||
| 7mb0 | 1.54 Å | 2022-10-12 | X-ray | YVG | SARS-CoV-2 | NSP5 (3CLpro) | 17.92% | 19.93% | 12.10% | D-phenylalanyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | D-phenylalanyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | 99.70% | 11.4 | NSLS-II (17-ID-2) | PHENIX | 2021-03-31 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 75.2 | - | No | - | - | 82.2 | 61.8 | 84.4 | - | |||||||
| 7mb1 | 1.43 Å | 2022-10-12 | X-ray | YVJ | SARS-CoV-2 | NSP5 (3CLpro) | 18.77% | 21.38% | 3.30% | 4-fluoro-D-phenylalanyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | 4-fluoro-D-phenylalanyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | 94.00% | 11.0 | NSLS-II (17-ID-2) | PHENIX | 2021-03-31 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 59.5 | - | No | - | - | 70.9 | 42.5 | 80.1 | - | |||||||
| 7mb2 | 1.89 Å | 2022-10-12 | X-ray | YVM | SARS-CoV-2 | NSP5 (3CLpro) | 16.66% | 21.04% | 11.00% | 4-fluoro-N-{3-[(prop-2-yn-1-yl)oxy]propanoyl}-D-phenylalanyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | 4-fluoro-N-{3-[(prop-2-yn-1-yl)oxy]propanoyl}-D-phenylalanyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | 99.50% | 8.0 | NSLS-II (17-ID-2) | PHENIX | 2021-03-31 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 80.8 | - | No | - | - | 73.7 | 86.1 | 85.7 | - | |||||||
| 7mb3 | 1.81 Å | 2022-10-12 | X-ray | YVV | SARS-CoV-2 | NSP5 (3CLpro) | 19.47% | 23.32% | 6.90% | N-{3-[(prop-2-yn-1-yl)oxy]propanoyl}-D-phenylalanyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | N-{3-[(prop-2-yn-1-yl)oxy]propanoyl}-D-phenylalanyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | 96.40% | 10.2 | NSLS-II (17-ID-2) | PHENIX | 2021-03-31 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 62.9 | - | No | - | - | 51.8 | 70.3 | 78.5 | - | |||||||
| 7zce | 3.50 Å | 2022-10-12 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-03-28 | 36114671 | - | - | - | - | N/A | - | - | scFv76 single chain fragment; Spike glycoprotein,Fibritin | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7zcf | 4.00 Å | 2022-10-12 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-03-28 | 36114671 | - | - | - | - | N/A | - | - | Spike glycoprotein,Fibritin; scFv76 single chain fragment | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8a4q | 1.75 Å | 2022-10-12 | X-ray | O6K | SARS-CoV-2 | NSP5 (3CLpro) | 17.80% | 21.92% | 10.00% | ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | 98.10% | 11.1 | EMBL/DESY, HAMBURG (X11) | REFMAC | 2022-06-13 | 36179320 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 61.2 | - | No | - | 65.8 | 64.4 | 66.6 | - | ||||||||
| 8a4t | 2.50 Å | 2022-10-12 | X-ray | O6K | SARS-CoV-2 | NSP5 (3CLpro) | 20.75% | 28.94% | 14.50% | ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | 100.00% | 10.7 | PETRA III, DESY (P11) | REFMAC | 2022-06-13 | 36179320 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 19.2 | - | No | - | 10.1 | 76.2 | 26.4 | - | ||||||||
| 8e25 | 1.87 Å | 2022-10-12 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | 19.40% | 24.00% | N/A | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | autoPROC | 51.42% | 7.4 | LNLS SIRUS (MANACA) | REFMAC | 2022-08-14 | 36775130 | - | - | - | - | N/A | - | - | Replicase polyprotein 1ab | 46.1 | - | No | - | 44.7 | 45.2 | 77.6 | - | |||||||
| 8e26 | 1.84 Å | 2022-10-12 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | 22.42% | 26.54% | N/A | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | autoPROC | 39.62% | 4.4 | LNLS SIRUS (MANACA) | REFMAC | 2022-08-14 | 36775130 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 35.5 | - | No | - | 22.7 | 71.3 | 53.0 | - | |||||||
| 8gx9 | 4.01 Å | 2022-10-12 | X-ray | NAG | SARS-CoV-2 | Spike | 23.78% | 29.69% | 29.00% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 97.27% | 4.8 | SSRF (BL17U1) | PHENIX | 2022-09-19 | - | - | - | - | - | N/A | - | - | light chain of P2B-1G5; heavy chain of P2C-1F11; light chain of P2C-1F11; Spike protein S1; heavy chain of P2B-1G5 | 23.9 | - | No | - | - | 8.0 | 73.1 | 41.9 | - | ||||||
| 7ufk | 2.38 Å | 2022-10-19 | X-ray | NAG, NA | SARS-CoV-2 | Spike | 19.19% | 23.53% | 4.30% | SODIUM ION; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose; SODIUM ION | XDS | 51.40% | 17.0 | SSRL (BL12-1) | PHENIX | 2022-03-22 | 36256797 | NA; ZN | - | - | - | N/A | - | - | Spike protein S1 - receptor-binding domain; Angiotensin-converting enzyme 2 - UNP residues 19-615 | 36.4 | - | No | - | 49.6 | 28.1 | 71.3 | - | |||||||
| 7ufl | 2.84 Å | 2022-10-19 | X-ray | NAG | SARS-CoV-2 | Spike | 21.37% | 27.09% | 6.10% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 54.50% | 10.0 | SSRL (BL12-1) | PHENIX | 2022-03-22 | 36256797 | ZN | - | - | - | N/A | - | - | Spike protein S1 - receptor-binding domain; Angiotensin-converting enzyme 2 | 5.5 | - | No | - | 19.1 | 18.2 | 34.5 | - | |||||||
| 7vhj | 4.20 Å | 2022-10-19 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-09-22 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7vhk | 3.60 Å | 2022-10-19 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-09-22 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7vhl | 3.90 Å | 2022-10-19 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-09-22 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7vhm | 3.20 Å | 2022-10-19 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-09-22 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7wse | 2.93 Å | 2022-10-19 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-01-29 | 36187898 | ZN | - | - | - | N/A | - | - | Spike protein S1; Angiotensin-converting enzyme | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7wsf | 2.87 Å | 2022-10-19 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-01-29 | 36187898 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme; Spike protein S1 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7ye7 | 2.95 Å | 2022-10-19 | X-ray | DD9, OCT | SARS-CoV-2 | ORF9B | 26.57% | 28.92% | 6.10% | nonane; N-OCTANE | nonane; N-OCTANE | HKL-2000 | 99.80% | 34.357 | SSRF (BL17U1) | PHENIX | 2022-07-05 | 36184694 | - | - | - | - | N/A | - | - | ORF9b protein | 5.2 | - | No | - | 10.2 | 23.5 | 37.1 | - | |||||||
| 7ye8 | 3.01 Å | 2022-10-19 | X-ray | OCT | SARS-CoV-2 | ORF9B | 24.16% | 29.85% | 6.50% | N-OCTANE | N-OCTANE | HKL-2000 | 100.00% | 46.615 | SSRF (BL17U1) | PHENIX | 2022-07-05 | 36184694 | - | - | - | - | N/A | - | - | ORF9b protein | 13.9 | - | No | - | 7.6 | 31.0 | 61.1 | - | |||||||
| 7yqt | 3.45 Å | 2022-10-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-08-08 | 36270286 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7yqu | 3.19 Å | 2022-10-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-08-08 | 36270286 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7yqv | 3.58 Å | 2022-10-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-08-08 | 36270286 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7yqw | 3.51 Å | 2022-10-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-08-08 | 36270286 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7yqx | 3.72 Å | 2022-10-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-08-08 | 36270286 | - | - | - | - | N/A | - | - | Spike glycoprotein; S309 light chain; S309 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7yqy | 3.74 Å | 2022-10-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-08-08 | 36270286 | - | - | - | - | N/A | - | - | S309 heavy chain; Spike glycoprotein; S309 light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7yqz | 3.84 Å | 2022-10-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-08-08 | 36270286 | - | - | - | - | N/A | - | - | Spike glycoprotein; S309 light chain; S309 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7yr0 | 3.98 Å | 2022-10-19 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-08-08 | 36270286 | - | - | - | - | N/A | - | - | Spike protein S1; IGK@ protein; Heavy chain of S309 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7yr1 | 3.62 Å | 2022-10-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-08-08 | 36270286 | - | - | - | - | N/A | - | - | Spike glycoprotein; XG2v024 Heavy chain; XG2v024 Light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7yr2 | 3.30 Å | 2022-10-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-08-08 | 36270286 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7yr3 | 3.52 Å | 2022-10-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-08-08 | 36270286 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8d8q | 4.20 Å | 2022-10-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-06-08 | 36202799 | - | - | - | - | N/A | - | - | 2196 heavy chain; 2196 light chain; Spike glycoprotein; 2130 light chain; 2130 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8d8r | 4.10 Å | 2022-10-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-06-08 | 36202799 | - | - | - | - | N/A | - | - | 2196 light chain; Spike glycoprotein; 2196 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8h10 | 2.99 Å | 2022-10-19 | Cryo-EM | BLA, NAG, EIC | SARS-CoV | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; LINOLEIC ACID; BILIVERDINE IX ALPHA | BILIVERDINE IX ALPHA; 2-acetamido-2-deoxy-beta-D-glucopyranose; LINOLEIC ACID | - | - | () | PHENIX | 2022-09-30 | 37402591 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8h13 | 4.05 Å | 2022-10-19 | Cryo-EM | NAG | SARS-CoV | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-09-30 | 37402591 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8h14 | 3.39 Å | 2022-10-19 | Cryo-EM | NAG, EIC | SARS-CoV | Spike | N/A | N/A | N/A | LINOLEIC ACID; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose; LINOLEIC ACID | - | - | () | PHENIX | 2022-09-30 | 37402591 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wb5 | 3.70 Å | 2022-10-26 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2021-12-15 | 35078968 | - | - | - | - | N/A | - | - | hu33 light chain; hu33 heavy chain; Surface glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 7wbh | 3.70 Å | 2022-10-26 | Cryo-EM | NAG | SARS-CoV | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-12-16 | 35078968 | - | - | - | - | N/A | - | - | heavy chain of hu33; Spike glycoprotein; Spike glycoprotein; light chain of hu33 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8erq | 3.30 Å | 2022-10-26 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-10-12 | 36264829 | - | - | - | - | N/A | - | - | S2X324 Fab heavy chain; S2X324 Fab light chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8err | 3.10 Å | 2022-10-26 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-10-12 | 36264829 | - | - | - | - | N/A | - | - | S2X324 light chain; S2X324 heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8gs6 | 2.86 Å | 2022-10-26 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-09-05 | 36272413 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7fr0 | 1.15 Å | 2022-11-09 | X-ray | WW0 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 12.69% | 14.51% | 6.40% | 2-hydroxy-N-(pentan-3-yl)-3H-imidazo[4,5-b]pyridine-7-carboxamide | 2-hydroxy-N-(pentan-3-yl)-3H-imidazo[4,5-b]pyridine-7-carboxamide | XDS | 99.70% | 14.37 | SSRL (BL12-1) | PHENIX | 2022-10-20 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 | 75.2 | - | No | - | - | 98.7 | 59.3 | 70.4 | - | |||||
| 7fr1 | 1.15 Å | 2022-11-09 | X-ray | WWC | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.23% | 15.18% | 5.70% | 1-methyl-N'-(7H-purin-6-yl)cyclopropane-1-carbohydrazide | 1-methyl-N'-(7H-purin-6-yl)cyclopropane-1-carbohydrazide | XDS | 99.20% | 14.21 | SSRL (BL12-1) | PHENIX | 2022-10-20 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 | 73.6 | - | No | - | - | 98.1 | 46.2 | 79.8 | - | |||||
| 7fr2 | 1.15 Å | 2022-11-09 | X-ray | WWH | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.25% | 15.30% | 4.80% | 3-cyclohexyl-N-{(2S)-1-[(9H-purin-6-yl)amino]butan-2-yl}propanamide | 3-cyclohexyl-N-{(2S)-1-[(9H-purin-6-yl)amino]butan-2-yl}propanamide | XDS | 99.10% | 18.93 | SSRL (BL12-1) | PHENIX | 2022-10-20 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 | 72.0 | - | No | - | - | 98.0 | 41.0 | 81.3 | - | |||||
| 7fr3 | 1.00 Å | 2022-11-09 | X-ray | WWH | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.56% | 16.64% | 11.20% | 3-cyclohexyl-N-{(2S)-1-[(9H-purin-6-yl)amino]butan-2-yl}propanamide | 3-cyclohexyl-N-{(2S)-1-[(9H-purin-6-yl)amino]butan-2-yl}propanamide | XDS | 99.10% | 6.57 | SSRL (BL12-1) | PHENIX | 2022-10-20 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 | 61.1 | - | No | - | - | 95.9 | 22.4 | 78.2 | - | |||||
| 7fr4 | 1.00 Å | 2022-11-09 | X-ray | WWN | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.81% | 15.76% | 9.00% | 2-cyclohexyl-N-{(2S)-1-[(7H-purin-6-yl)amino]butan-2-yl}acetamide | 2-cyclohexyl-N-{(2S)-1-[(7H-purin-6-yl)amino]butan-2-yl}acetamide | XDS | 99.80% | 9.74 | SSRL (BL12-1) | PHENIX | 2022-10-20 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 | 58.4 | - | No | - | - | 97.4 | 17.7 | 76.3 | - | |||||
| 7fr5 | 1.00 Å | 2022-11-09 | X-ray | WWT | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.87% | 15.58% | 10.80% | 2-hydroxy-N-propyl-3H-imidazo[4,5-b]pyridine-7-carboxamide | 2-hydroxy-N-propyl-3H-imidazo[4,5-b]pyridine-7-carboxamide | XDS | 99.70% | 8.3 | SSRL (BL12-1) | PHENIX | 2022-10-20 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 | 70.5 | - | No | - | - | 97.7 | 26.7 | 92.4 | - | |||||
| 7fr6 | 1.00 Å | 2022-11-09 | X-ray | WX6 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.42% | 16.25% | 7.60% | (azepan-1-yl)(2-hydroxy-3H-imidazo[4,5-b]pyridin-7-yl)methanone | (azepan-1-yl)(2-hydroxy-3H-imidazo[4,5-b]pyridin-7-yl)methanone | XDS | 99.80% | 10.46 | SSRL (BL12-1) | PHENIX | 2022-10-20 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 | 63.2 | - | No | - | - | 96.7 | 26.7 | 77.8 | - | |||||
| 7fr7 | 1.00 Å | 2022-11-09 | X-ray | WXB | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 15.85% | 17.35% | 12.00% | 6-(azepane-1-carbonyl)pyrido[2,3-d]pyrimidine-2,4(1H,3H)-dione | 6-(azepane-1-carbonyl)pyrido[2,3-d]pyrimidine-2,4(1H,3H)-dione | XDS | 99.60% | 7.33 | SSRL (BL12-1) | PHENIX | 2022-10-20 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 | 58.7 | - | No | - | - | 94.2 | 26.7 | 71.1 | - | |||||
| 7fr8 | 1.00 Å | 2022-11-09 | X-ray | WXF | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.99% | 15.58% | 12.10% | (2-hydroxy-3H-imidazo[4,5-b]pyridin-7-yl)[(2R)-2-methylmorpholin-4-yl]methanone | (2-hydroxy-3H-imidazo[4,5-b]pyridin-7-yl)[(2R)-2-methylmorpholin-4-yl]methanone | XDS | 99.40% | 7.71 | SSRL (BL12-1) | PHENIX | 2022-10-20 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 | 68.4 | - | No | - | - | 97.7 | 29.3 | 85.2 | - | |||||
| 7fr9 | 1.00 Å | 2022-11-09 | X-ray | WXK | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.61% | 16.57% | 8.80% | N-(1-methylcyclopropyl)-2,4-dioxo-1,2,3,4-tetrahydropyrido[2,3-d]pyrimidine-6-sulfonamide | N-(1-methylcyclopropyl)-2,4-dioxo-1,2,3,4-tetrahydropyrido[2,3-d]pyrimidine-6-sulfonamide | XDS | 99.80% | 8.49 | SSRL (BL12-1) | PHENIX | 2022-10-20 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 | 65.9 | - | No | - | - | 96.1 | 38.5 | 72.1 | - | |||||
| 7fra | 1.00 Å | 2022-11-09 | X-ray | WXO | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.36% | 15.93% | 14.00% | 6-(azepane-1-sulfonyl)pyrido[2,3-d]pyrimidine-2,4(1H,3H)-dione | 6-(azepane-1-sulfonyl)pyrido[2,3-d]pyrimidine-2,4(1H,3H)-dione | XDS | 99.50% | 5.92 | SSRL (BL12-1) | PHENIX | 2022-10-20 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 | 59.0 | - | No | - | - | 97.2 | 31.0 | 64.4 | - | |||||
| 7frb | 1.00 Å | 2022-11-09 | X-ray | WXT | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.92% | 15.36% | 12.50% | 1-methyl-N-{(2S)-3-methyl-2-[(9H-purin-6-yl)amino]butyl}cyclobutane-1-carboxamide | 1-methyl-N-{(2S)-3-methyl-2-[(9H-purin-6-yl)amino]butyl}cyclobutane-1-carboxamide | XDS | 99.70% | 7.31 | SSRL (BL12-1) | PHENIX | 2022-10-20 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 | 68.1 | - | No | - | - | 97.9 | 29.3 | 84.3 | - | |||||
| 7frc | 1.00 Å | 2022-11-09 | X-ray | WXX | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 13.98% | 15.84% | 6.80% | 3-cyclohexyl-N-{(2R)-2-[(1H-pyrazolo[3,4-d]pyrimidin-4-yl)amino]butyl}propanamide | 3-cyclohexyl-N-{(2R)-2-[(1H-pyrazolo[3,4-d]pyrimidin-4-yl)amino]butyl}propanamide | XDS | 99.50% | 11.33 | SSRL (BL12-1) | PHENIX | 2022-10-20 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 | 63.7 | - | No | - | - | 97.3 | 27.9 | 76.9 | - | |||||
| 7frd | 1.00 Å | 2022-11-09 | X-ray | WY6 | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.56% | 16.20% | 11.60% | 3-phenyl-N-{(2S)-1-[(7H-purin-6-yl)amino]butan-2-yl}propanamide | 3-phenyl-N-{(2S)-1-[(7H-purin-6-yl)amino]butan-2-yl}propanamide | XDS | 99.50% | 8.31 | SSRL (BL12-1) | PHENIX | 2022-10-20 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 | 53.1 | - | No | - | - | 96.8 | 17.7 | 66.6 | - | |||||
| 7sd9 | 1.85 Å | 2022-11-09 | X-ray | 8T6 | SARS-CoV-2 | NSP5 (3CLpro) | 22.22% | 24.94% | N/A | N-[(2S)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4,4-dimethyl-1-oxopentan-2-yl]-1H-indole-2-carboxamide | N-[(2S)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4,4-dimethyl-1-oxopentan-2-yl]-1H-indole-2-carboxamide | PROTEUM PLUS | 95.40% | 13.3 | ROTATING ANODE () | PHENIX | 2021-09-29 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 13.0 | - | No | - | - | 35.7 | 21.1 | 40.5 | - | ||||||
| 7sda | 1.85 Å | 2022-11-09 | X-ray | 8UI | SARS-CoV-2 | NSP5 (3CLpro) | 21.70% | 24.44% | N/A | N-[(2S)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4,4-dimethyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | N-[(2S)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4,4-dimethyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | PROTEUM PLUS | 99.70% | 10.4 | ROTATING ANODE () | PHENIX | 2021-09-29 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 19.1 | - | No | - | - | 40.7 | 16.8 | 55.0 | - | ||||||
| 7sdc | 1.85 Å | 2022-11-09 | X-ray | I80 | SARS-CoV-2 | NSP5 (3CLpro) | 27.56% | 30.02% | N/A | (1R,2S,5S)-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-{[4-(trifluoromethoxy)phenoxy]acetyl}-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-{[4-(trifluoromethoxy)phenoxy]acetyl}-3-azabicyclo[3.1.0]hexane-2-carboxamide | PROTEUM PLUS | 98.40% | 9.5 | ROTATING ANODE () | PHENIX | 2021-09-29 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 2.9 | - | No | - | - | 7.2 | 13.5 | 39.0 | - | ||||||
| 7wsg | 3.03 Å | 2022-11-09 | Cryo-EM | - | SARS-CoV | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-01-29 | 36187898 | ZN | - | - | - | N/A | - | - | Spike protein S1; Angiotensin-converting enzyme | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7wsh | 2.89 Å | 2022-11-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-29 | 36187898 | ZN | - | - | - | N/A | - | - | Spike protein S1; Angiotensin-converting enzyme | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7x29 | 2.49 Å | 2022-11-09 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2022-02-25 | 36246239 | - | - | - | - | N/A | - | - | antibody S41 heavy chain; antibody S41 light chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 7x2a | 2.49 Å | 2022-11-09 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2022-02-25 | 36246239 | - | - | - | - | N/A | - | - | antibody S41 heavy chain; MERS-CoV Spike glycoprotein; antibody S41 light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 7xar | 1.60 Å | 2022-11-09 | X-ray | BOV, NI | SARS-CoV-2 | NSP5 (3CLpro) | 17.32% | 20.64% | 5.90% | 4-fluoranyl-~{N}-[(2~{S})-1-[2-(2-fluoranylethanoyl)-2-[[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]methyl]hydrazinyl]-4-methyl-1-oxidanylidene-pentan-2-yl]-1~{H}-indole-2-carboxamide; NICKEL (II) ION | 4-fluoranyl-~{N}-[(2~{S})-1-[2-(2-fluoranylethanoyl)-2-[[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]methyl]hydrazinyl]-4-methyl-1-oxidanylidene-pentan-2-yl]-1~{H}-indole-2-carboxamide; NICKEL (II) ION | XDS | 99.50% | 18.3 | Photon Factory (BL-5A) | REFMAC | 2022-03-18 | 36229406 | NI | - | - | - | N/A | - | - | 3C-like proteinase | 69.6 | - | No | - | 77.0 | 54.2 | 83.5 | - | |||||||
| 7zr2 | 1.45 Å | 2022-11-09 | X-ray | - | SARS-CoV-2 | Spike | 17.40% | 20.90% | 5.90% | Spike protein S2' | - | XDS | 96.30% | 9.4 | ALBA (XALOC) | PHENIX | 2022-05-03 | 36220405 | - | - | - | - | N/A | - | - | Spike protein S2'; Spike protein S2',Chimeric protein mimic of SARS-CoV-2 Spike HR1 | - | - | No | - | 74.8 | - | - | - | |||||||
| 8ers | 1.05 Å | 2022-11-09 | X-ray | WQO | SARS-CoV-2 | NSP3: Macro | PanDDA fragment screening | 14.22% | 16.24% | 3.00% | (1R,2S)-1-[(4-amino-2-hydroxybenzoyl)oxy]-2,3-dihydro-1H-indene-2-carboxylic acid | (1R,2S)-1-[(4-amino-2-hydroxybenzoyl)oxy]-2,3-dihydro-1H-indene-2-carboxylic acid | XDS | 99.80% | 25.06 | ALS (8.3.1) | PHENIX | 2022-10-12 | - | - | - | - | - | N/A | - | - | Non-structural protein 3 - macrodomain | 72.5 | - | No | - | - | 96.7 | 52.3 | 72.5 | - | |||||
| 8h0x | 2.57 Å | 2022-11-09 | Cryo-EM | BLA, EIC, NAG | SARS-CoV | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; LINOLEIC ACID; BILIVERDINE IX ALPHA | BILIVERDINE IX ALPHA; LINOLEIC ACID; 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-09-30 | 37402591 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8h0y | 2.85 Å | 2022-11-09 | Cryo-EM | NAG, BLA, EIC | SARS-CoV | Spike | N/A | N/A | N/A | LINOLEIC ACID; 2-acetamido-2-deoxy-beta-D-glucopyranose; BILIVERDINE IX ALPHA | 2-acetamido-2-deoxy-beta-D-glucopyranose; BILIVERDINE IX ALPHA; LINOLEIC ACID | - | - | () | PHENIX | 2022-09-30 | 37402591 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8h0z | 2.99 Å | 2022-11-09 | Cryo-EM | NAG, BLA, EIC | SARS-CoV | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; BILIVERDINE IX ALPHA; LINOLEIC ACID | 2-acetamido-2-deoxy-beta-D-glucopyranose; BILIVERDINE IX ALPHA; LINOLEIC ACID | - | - | () | PHENIX | 2022-09-30 | 37402591 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8h11 | 2.72 Å | 2022-11-09 | Cryo-EM | NAG | SARS-CoV | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-09-30 | 37402591 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8h12 | 3.45 Å | 2022-11-09 | Cryo-EM | NAG | SARS-CoV | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-09-30 | 37402591 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8h15 | 3.14 Å | 2022-11-09 | Cryo-EM | NAG | SARS-CoV | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-09-30 | 37402591 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8h16 | 3.36 Å | 2022-11-09 | Cryo-EM | NAG | SARS-CoV | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-09-30 | 37402591 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7q3q | 2.30 Å | 2022-11-16 | X-ray | - | SARS-CoV-2 | Spike | 18.49% | 22.02% | 11.50% | - | XDS | 99.40% | 32.4 | SOLEIL (PROXIMA 2) | PHENIX | 2021-10-28 | - | - | - | - | - | N/A | - | - | VHH-12; Spike glycoprotein | 83.4 | - | No | - | - | 64.8 | 93.5 | 98.0 | - | |||||||
| 7q3r | 1.92 Å | 2022-11-16 | X-ray | - | SARS-CoV-2 | Spike | 18.19% | 20.90% | 7.90% | - | XDS | 99.80% | 22.5 | SOLEIL (PROXIMA 2) | PHENIX | 2021-10-28 | - | - | - | - | - | N/A | - | - | VHH-F04; VHH-G09; Spike glycoprotein | 64.4 | - | No | - | - | 74.8 | 38.5 | 90.3 | - | |||||||
| 7six | 3.16 Å | 2022-11-16 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX; phenix.real_space_refine | 2021-10-15 | - | - | - | - | - | N/A | - | - | N3-1 Fab heavy chain; N3-1 Fab light chain; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7sj0 | 3.36 Å | 2022-11-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2021-10-15 | - | - | - | - | - | N/A | - | - | A7V3 Fab heavy chain; A7V3 Fab light chain; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 8eyj | 1.74 Å | 2022-11-16 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | 19.31% | 23.39% | 7.00% | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | autoPROC | 71.50% | 8.9 | LNLS SIRUS (MANACA) | REFMAC | 2022-10-27 | 36941262 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 55.0 | - | No | - | 51.3 | 71.0 | 62.5 | - | |||||||
| 7q5e | 1.67 Å | 2022-11-23 | X-ray | 90I, NA | SARS-CoV-2 | NSP5 (3CLpro) | 17.07% | 19.36% | 7.20% | SODIUM ION; benzyl (S)-2-(((S)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)-4-(phenethylamino)butan-2-yl)carbamoyl)pyrrolidine-1-carboxylate | benzyl (S)-2-(((S)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)-4-(phenethylamino)butan-2-yl)carbamoyl)pyrrolidine-1-carboxylate; SODIUM ION | XDS | 100.00% | 16.2 | ELETTRA (11.2C) | PHENIX; PHENIX | 2021-11-03 | 36332546 | - | - | - | - | N/A | - | - | 3C-like proteinase | 75.9 | - | No | - | 85.8 | 50.5 | 93.9 | - | |||||||
| 7q5f | 1.72 Å | 2022-11-23 | X-ray | 90X, NA, NO3 | SARS-CoV-2 | NSP5 (3CLpro) | 17.24% | 19.58% | 10.40% | NITRATE ION; SODIUM ION; (S)-1-(2-(2,4-dichlorophenoxy)acetyl)-N-((S)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)-4-(phenethylamino)butan-2-yl)pyrrolidine-2-carboxamide | (S)-1-(2-(2,4-dichlorophenoxy)acetyl)-N-((S)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)-4-(phenethylamino)butan-2-yl)pyrrolidine-2-carboxamide; SODIUM ION; NITRATE ION | XDS | 100.00% | 14.0 | ELETTRA (11.2C) | PHENIX; PHENIX | 2021-11-03 | 36332546 | - | - | - | - | N/A | - | - | 3C-like proteinase | 68.7 | - | No | - | 84.5 | 48.8 | 79.5 | - | |||||||
| 7s83 | 2.52 Å | 2022-11-23 | X-ray | - | SARS-CoV-2 | Spike | 23.29% | 27.81% | N/A | - | XDS | 99.49% | 11.57 | APS (24-ID-E) | PHENIX | 2021-09-17 | 36737435 | - | - | - | - | N/A | - | - | ShAb01 VNAR; ShAb02 VNAR; Spike protein S1 - receptor binding domain (RBD) | 5.0 | - | No | - | 15.0 | 21.0 | 33.8 | - | ||||||||
| 7sww | 3.13 Å | 2022-11-23 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-21 | - | - | - | - | - | N/A | - | - | SARS2-57 Fv light chain; Spike protein S1 - N-terminal domain (UNP residues 14-304); SARS2-57 Fv heavy chain | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7swx | 3.13 Å | 2022-11-23 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-11-21 | - | - | - | - | - | N/A | - | - | SARS2-57 Fv heavy chain; SARS2-57 Fv light chain; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7u9o | 3.40 Å | 2022-11-23 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-03-11 | 36302375 | - | - | - | - | N/A | - | - | NE12 Fab heavy chain; NE12 Fab light chain; Spike glycoprotein - Receptor-binding domain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7u9p | 3.50 Å | 2022-11-23 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-03-11 | 36302375 | - | - | - | - | N/A | - | - | NA8 Fab light chain; Spike glycoprotein - Receptor-binding domain; NA8 Fab heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wxz | 2.41 Å | 2022-11-23 | X-ray | - | SARS-CoV-2 | Spike | 24.90% | 29.37% | N/A | - | xia2 | 99.00% | 10.62 | SSRF (BL02U1) | PHENIX | 2022-02-15 | 36357786 | - | - | - | - | N/A | - | - | Spike protein S2' | 2.3 | - | No | - | 8.9 | 23.9 | 22.6 | - | ||||||||
| 7xc3 | 1.70 Å | 2022-11-23 | X-ray | - | SARS-CoV-2 | NSP3(SUD-M) | 19.63% | 21.79% | 15.40% | - | HKL-2000 | 99.30% | 29.0 | SSRF (BL18U1) | PHENIX | 2022-03-22 | 36356292 | - | - | - | - | N/A | - | - | Papain-like protease nsp3 | 84.5 | - | No | - | 67.2 | 95.7 | 99.4 | - | ||||||||
| 7xc4 | 2.10 Å | 2022-11-23 | X-ray | BJ6 | SARS-CoV-2 | NSP3(SUD-M) | 17.94% | 21.39% | N/A | 3-(4,5-diphenyl-1,3-oxazol-2-yl)propanoic acid | 3-(4,5-diphenyl-1,3-oxazol-2-yl)propanoic acid | HKL-2000 | 99.50% | 24.0 | SSRF (BL18U1) | PHENIX | 2022-03-22 | 36356292 | - | - | - | - | N/A | - | - | Papain-like protease nsp3 | 79.9 | - | No | - | 70.8 | 74.7 | 97.1 | - | |||||||
| 7xmx | 3.62 Å | 2022-11-23 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-04-27 | 36075908 | - | - | - | - | N/A | - | - | F61 heavy chain; F61 light chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7xmz | 3.25 Å | 2022-11-23 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-04-27 | 36075908 | - | - | - | - | N/A | - | - | D2 heavy chain; Spike glycoprotein; D2 light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7xst | 3.04 Å | 2022-11-23 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-05-15 | 36075908 | - | - | - | - | N/A | - | - | D2 heavy chain; Spike glycoprotein; F61 heavy chain; F61 light chain; D2 light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8a4y | 1.10 Å | 2022-11-23 | X-ray | QO6 | SARS-CoV-2 | NSP1 | 14.68% | 16.64% | N/A | N-(2,3-dihydro-1H-inden-5-yl)acetamide | N-(2,3-dihydro-1H-inden-5-yl)acetamide | autoPROC | 99.90% | 18.4 | ESRF (MASSIF-1) | PHENIX | 2022-06-13 | 36412088 | - | - | - | - | N/A | - | - | Host translation inhibitor nsp1 | 57.3 | - | No | - | 95.9 | 23.7 | 69.5 | - | |||||||
| 8a55 | 0.99 Å | 2022-11-23 | X-ray | - | SARS-CoV-2 | NSP1 | 15.34% | 17.34% | N/A | - | XDS | 99.49% | 12.73 | ESRF (ID30B) | PHENIX | 2022-06-14 | 36293303 | - | - | - | - | N/A | - | - | Host translation inhibitor nsp1 | 43.0 | - | No | - | 94.3 | 26.9 | 40.5 | - | ||||||||
| 8ays | 1.37 Å | 2022-11-23 | X-ray | 92G | SARS-CoV-2 | NSP1 | 14.29% | 15.00% | N/A | 4-(2-amino-1,3-thiazol-4-yl)phenol | 4-(2-amino-1,3-thiazol-4-yl)phenol | DIALS | 93.70% | 23.7 | ESRF (MASSIF-1) | PHENIX | 2022-09-03 | 36293303 | - | - | - | - | N/A | - | - | Host translation inhibitor nsp1 | 53.5 | - | No | - | 98.3 | 42.6 | 40.9 | - | |||||||
| 8az8 | 1.18 Å | 2022-11-23 | X-ray | OEI | SARS-CoV-2 | NSP1 | 16.37% | 20.69% | N/A | 2-[(phenylmethyl)amino]ethanol | 2-[(phenylmethyl)amino]ethanol | XDS | 92.80% | 18.3 | ESRF (MASSIF-1) | PHENIX | 2022-09-05 | 36293303 | - | - | - | - | N/A | - | - | Host translation inhibitor nsp1 | 51.1 | - | No | - | 76.7 | 23.5 | 77.0 | - | |||||||
| 8bh5 | 2.38 Å | 2022-11-23 | X-ray | PO4 | SARS-CoV-2 | Spike | 18.57% | 23.25% | 24.00% | PHOSPHATE ION | PHOSPHATE ION | xia2 | 94.80% | 6.3 | Diamond (I03) | PHENIX | 2022-10-29 | 36319620 | - | - | - | - | N/A | - | - | Spike protein S1; Nanobody C1; Beta-27 heavy chain; Beta-27 light chain | 39.2 | - | No | - | 52.6 | 14.9 | 86.9 | - | |||||||
| 8e4j | 1.90 Å | 2022-11-23 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 16.95% | 20.68% | 8.50% | - | CrysalisPro | 95.80% | 16.02 | ROTATING ANODE () | PHENIX | 2022-08-18 | 36334779 | - | - | - | - | N/A | - | - | Replicase polyprotein 1ab | 45.5 | - | No | - | 76.8 | 24.1 | 65.6 | - | ||||||||
| 8e4r | 1.80 Å | 2022-11-23 | X-ray | K36 | SARS-CoV-2 | NSP5 (3CLpro) | 16.48% | 19.50% | 6.60% | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | CrysalisPro | 99.20% | 21.18 | ROTATING ANODE () | PHENIX | 2022-08-18 | 36334779 | - | - | - | - | N/A | - | - | Replicase polyprotein 1ab | 63.5 | - | No | - | 84.9 | 46.7 | 70.0 | - | |||||||
| 8f44 | 1.65 Å | 2022-11-23 | X-ray | XFR, PG4, XFF | SARS-CoV-2 | NSP5 (3CLpro) | 18.10% | 21.90% | 6.20% | (1R,2S)-1-hydroxy-2-[(N-{[2-methyl-2-(phenylsulfanyl)propoxy]carbonyl}-L-leucyl)amino]-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; (2-methyl-2-phenylsulfanyl-propyl) ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | (1R,2S)-1-hydroxy-2-[(N-{[2-methyl-2-(phenylsulfanyl)propoxy]carbonyl}-L-leucyl)amino]-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; TETRAETHYLENE GLYCOL; (2-methyl-2-phenylsulfanyl-propyl) ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | XDS | 99.90% | 12.2 | NSLS-II (19-ID) | PHENIX | 2022-11-10 | - | - | - | - | - | N/A | - | - | 3C-like proteinase - Full Length | 73.2 | - | No | - | - | 65.9 | 78.4 | 79.1 | - | ||||||
| 8f45 | 1.65 Å | 2022-11-23 | X-ray | XF8 | SARS-CoV-2 | NSP5 (3CLpro) | 17.17% | 20.37% | 6.40% | (2-methyl-2-phenylsulfanyl-propyl) ~{N}-[(2~{S})-1-[[(2~{S},3~{S})-3-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | (2-methyl-2-phenylsulfanyl-propyl) ~{N}-[(2~{S})-1-[[(2~{S},3~{S})-3-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | XDS | 100.00% | 30.6 | SEALED TUBE () | PHENIX | 2022-11-10 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 75.5 | - | No | - | - | 79.1 | 63.6 | 86.3 | - | ||||||
| 8f46 | 1.50 Å | 2022-11-23 | X-ray | XCK, PG4 | SARS-CoV-2 | NSP5 (3CLpro) | 18.54% | 22.08% | 4.90% | N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-{[2-methyl-2-(phenylsulfanyl)propoxy]carbonyl}-L-leucinamide | N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-{[2-methyl-2-(phenylsulfanyl)propoxy]carbonyl}-L-leucinamide; TETRAETHYLENE GLYCOL | XDS | 99.80% | 14.2 | NSLS-II (19-ID) | PHENIX | 2022-11-10 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | 73.1 | - | No | - | - | 64.4 | 78.4 | 80.4 | - | ||||||
| 7u6r | 2.50 Å | 2022-11-30 | Cryo-EM | NAG | PDF-2180 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-03-05 | 36477529 | - | - | - | - | N/A | - | - | PDF-2180 Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 7uo4 | 3.38 Å | 2022-11-30 | Cryo-EM | NWX | SARS-CoV-2 | NSP7/NSP8/NSP12 | N/A | N/A | N/A | [[(2~{R},3~{S},4~{R},5~{R})-5-(4-azanylpyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-cyano-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate; Product RNA (35-MER); Template RNA (55-MER) | [[(2~{R},3~{S},4~{R},5~{R})-5-(4-azanylpyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-cyano-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | - | - | () | PHENIX | 2022-04-12 | 36725929 | ZN; MG | - | - | - | N/A | - | - | Non-structural protein 7; RNA-directed RNA polymerase - UNP residues 4393-5324; Product RNA (35-MER); Template RNA (55-MER); Non-structural protein 8 | N/A | - | No | - | N/A | N/A | N/A | Template RNA (55-MER); Product RNA (35-MER) | - | |||||||
| 7uo7 | 3.09 Å | 2022-11-30 | Cryo-EM | ATP | SARS-CoV-2 | NSP7/NSP8/NSP12 | N/A | N/A | N/A | ADENOSINE-5'-TRIPHOSPHATE; Template RNA (55-MER); Product RNA (35-MER) | ADENOSINE-5'-TRIPHOSPHATE | - | - | () | PHENIX | 2022-04-12 | 36725929 | ZN; MG | - | - | - | N/A | - | - | Template RNA (55-MER); Product RNA (35-MER); Non-structural protein 7; Non-structural protein 8; RNA-directed RNA polymerase - UNP residues 4393-5324 | N/A | - | No | - | N/A | N/A | N/A | Template RNA (55-MER); Product RNA (35-MER) | - | |||||||
| 7uo9 | 3.13 Å | 2022-11-30 | Cryo-EM | UTP | SARS-CoV-2 | NSP7/NSP8/NSP12 | N/A | N/A | N/A | Product RNA (35-MER); URIDINE 5'-TRIPHOSPHATE; Template RNA (55-MER) | URIDINE 5'-TRIPHOSPHATE | - | - | () | PHENIX | 2022-04-12 | 36725929 | MG; ZN | - | - | - | N/A | - | - | Non-structural protein 8; Non-structural protein 7; Product RNA (35-MER); Template RNA (55-MER); RNA-directed RNA polymerase - UNP residues 4393-5324 | N/A | - | No | - | N/A | N/A | N/A | Product RNA (35-MER); Template RNA (55-MER) | - | |||||||
| 7uob | 2.68 Å | 2022-11-30 | Cryo-EM | L2B, GTP | SARS-CoV-2 | NSP7/NSP8/NSP12 | N/A | N/A | N/A | 3'-DEOXYURIDINE-5'-MONOPHOSPHATE; Product RNA (35-MER); Template RNA (55-MER); GUANOSINE-5'-TRIPHOSPHATE | 3'-DEOXYURIDINE-5'-MONOPHOSPHATE; GUANOSINE-5'-TRIPHOSPHATE | - | - | () | PHENIX | 2022-04-12 | 36725929 | ZN; MG | - | - | - | N/A | - | - | Template RNA (55-MER); RNA-directed RNA polymerase - UNP residues 4393-5324; Non-structural protein 7; Non-structural protein 8; Product RNA (35-MER) | N/A | - | No | - | N/A | N/A | N/A | Product RNA (35-MER); Template RNA (55-MER) | - | |||||||
| 7uoe | 2.67 Å | 2022-11-30 | Cryo-EM | L2B, CTP | SARS-CoV-2 | NSP7/NSP8/NSP12 | N/A | N/A | N/A | 3'-DEOXYURIDINE-5'-MONOPHOSPHATE; Template RNA (55-MER); Product RNA (35-MER); CYTIDINE-5'-TRIPHOSPHATE | 3'-DEOXYURIDINE-5'-MONOPHOSPHATE; CYTIDINE-5'-TRIPHOSPHATE | - | - | () | PHENIX | 2022-04-12 | 36725929 | ZN; MG | - | - | - | N/A | - | - | Non-structural protein 7; Non-structural protein 8; RNA-directed RNA polymerase - UNP residues 4393-5324; Template RNA (55-MER); Product RNA (35-MER) | N/A | - | No | - | N/A | N/A | N/A | Template RNA (55-MER); Product RNA (35-MER) | - | |||||||
| 7wop | 3.51 Å | 2022-11-30 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-01-22 | 36207299 | - | - | - | - | N/A | - | - | GW01; Spike protein S1 - RBD; 16L9 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7woq | 3.47 Å | 2022-11-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-22 | 36207299 | - | - | - | - | N/A | - | - | Spike glycoprotein; 16L9 Fv | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wor | 3.70 Å | 2022-11-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-22 | 36207299 | - | - | - | - | N/A | - | - | 16L9 Fv; Spike glycoprotein; GW01 Fv | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wos | 3.91 Å | 2022-11-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-22 | 36207299 | - | - | - | - | N/A | - | - | Spike glycoprotein; 16L9 Fv; GW01 Fv | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wou | 3.47 Å | 2022-11-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-22 | 36207299 | - | - | - | - | N/A | - | - | Spike glycoprotein; 16L9 Fv; GW01 Fv | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wov | 3.87 Å | 2022-11-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-22 | 36207299 | - | - | - | - | N/A | - | - | GW01 Fv; Spike glycoprotein; 16L9 Fv | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wow | 6.11 Å | 2022-11-30 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-01-22 | 36207299 | - | - | - | - | N/A | - | - | Spike glycoprotein; GW01 Fv; 16L9 Fv | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7y75 | 3.10 Å | 2022-11-30 | Cryo-EM | 3PH, NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE; 2-acetamido-2-deoxy-beta-D-glucopyranose | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE; 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-06-21 | 36384914 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Sodium- and chloride-dependent transporter XTRP3; Spike protein S1 - BA.2 RBD | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7zv5 | 2.00 Å | 2022-11-30 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 20.14% | 23.98% | N/A | inhibitor TRIP5 | - | autoPROC | 82.20% | 14.4 | ESRF (MASSIF-1) | BUSTER | 2022-05-13 | 36332548 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5; inhibitor TRIP5 | 40.0 | - | No | - | 45.2 | 15.8 | 94.8 | - | |||||||
| 8dya | 3.67 Å | 2022-11-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-08-03 | 36356052 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8gwo | 3.80 Å | 2022-11-30 | Cryo-EM | GNP | SARS-CoV-2 | NSP7/NSP8/NSP9/NSP12/NSP13 | N/A | N/A | N/A | template; PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER; RNA (25-MER) | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | - | - | () | PHENIX | 2022-09-17 | 36335936 | ZN | - | - | - | N/A | - | - | RNA (25-MER); template; Helicase - UNP residues 5325-5925; Non-structural protein 9; Non-structural protein 8 - UNP residues 3943-4140; RNA-directed RNA polymerase - UNP residues 4393-5324; Non-structural protein 7 - UNP residues 3860-3942 | N/A | - | No | - | N/A | N/A | N/A | RNA (25-MER) | - | |||||||
| 7lcr | 1.95 Å | 2022-12-07 | X-ray | XTM | SARS-CoV-2 | NSP5 (3CLpro) | 23.84% | 27.67% | N/A | N~2~-{[(3-fluorophenyl)methoxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | N~2~-{[(3-fluorophenyl)methoxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | XDS | 99.82% | 8.1 | SSRL (BL12-2) | PHENIX | 2021-01-11 | 34118724 | - | - | - | - | N/A | - | - | 3C-like proteinase | 2.2 | - | No | - | 15.8 | 12.1 | 27.1 | - | |||||||
| 7tyz | 3.51 Å | 2022-12-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | DARPin FSR22; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-15 | 36411391 | - | - | - | - | N/A | - | - | Spike glycoprotein; DARPin FSR22 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7tz0 | 4.17 Å | 2022-12-07 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | DARPin FSR22 | - | - | - | () | PHENIX | 2022-02-15 | 36411391 | - | - | - | - | N/A | - | - | DARPin FSR22; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7uz4 | 3.10 Å | 2022-12-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-05-08 | 36370711 | - | - | - | - | N/A | - | - | M8a-3 Fab heavy chain; Spike glycoprotein - Spike 6P; M8a-3 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7uz5 | 3.20 Å | 2022-12-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-05-08 | 36370711 | - | - | - | - | N/A | - | - | M8a-6 Fab heavy chain; M8a-6 Fab light chain; Spike glycoprotein - Spike 6P | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7uz6 | 2.80 Å | 2022-12-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-05-08 | 36370711 | - | - | - | - | N/A | - | - | M8a-28 Fab heavy chain; Spike glycoprotein - Spike 6P; M8a-28 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7uz7 | 2.90 Å | 2022-12-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-05-08 | 36370711 | - | - | - | - | N/A | - | - | M8a-31 Fab light chain; M8a-31 Fab heavy chain; Spike glycoprotein - Spike 6P | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7uz8 | 3.10 Å | 2022-12-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-05-08 | 36370711 | - | - | - | - | N/A | - | - | M8a-31 Fab heavy chain; Spike glycoprotein - Omicron BA.1 Spike 6P; M8a-31 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7uz9 | 3.50 Å | 2022-12-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-05-08 | 36370711 | - | - | - | - | N/A | - | - | Spike glycoprotein - Spike 6P; M8a-34 Fab heavy chain; M8a-34 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7uza | 3.10 Å | 2022-12-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-05-08 | 36370711 | - | - | - | - | N/A | - | - | Spike glycoprotein - Spike 6P; HSW-1 Fab light chain; HSW-1 Fab heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7uzb | 4.10 Å | 2022-12-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-05-08 | 36370711 | - | - | - | - | N/A | - | - | Spike glycoprotein - Spike S1 domain; HSW-2 Fab light chain; HSW-2 Fab heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7vhn | 3.80 Å | 2022-12-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-09-22 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7wvl | 3.00 Å | 2022-12-07 | X-ray | - | SARS-CoV-2 | Spike | 23.26% | 27.94% | 9.30% | - | iMOSFLM | 100.00% | 7.3 | ESRF (MASSIF-1) | PHENIX | 2022-02-10 | 36508467 | - | - | - | - | N/A | - | - | P4A2 Fab heavy chain; P4A2 Fab Light Chain; Spike protein S1 - Receptor binding domain | 23.1 | - | No | - | 14.3 | 83.5 | 23.5 | - | ||||||||
| 7x2l | 2.40 Å | 2022-12-07 | X-ray | - | SARS-CoV-2 | Spike | 23.01% | 28.70% | N/A | - | HKL-2000 | 97.37% | 23.43 | SSRF (BL02U1) | PHENIX | 2022-02-25 | 36575191 | - | - | - | - | N/A | - | - | Spike protein S1 - RBD; Nanobody 3-2A2-4 | 13.1 | - | No | - | 11.0 | 23.8 | 62.7 | - | ||||||||
| 7x8w | 3.10 Å | 2022-12-07 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX; phenix.real_space_refine | 2022-03-15 | 36406861 | - | - | - | - | N/A | - | - | Ab354 heavy chain; Spike glycoprotein; Ab354 light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7x8y | 4.10 Å | 2022-12-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-03-15 | 36406861 | - | - | - | - | N/A | - | - | Ab159 light chain; Ab159 heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7x8z | 4.10 Å | 2022-12-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-03-15 | 36406861 | - | - | - | - | N/A | - | - | Ab188 heavy chain; Spike glycoprotein; Ab188 light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7x90 | 4.20 Å | 2022-12-07 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX; phenix.real_space_refine | 2022-03-15 | 36406861 | - | - | - | - | N/A | - | - | Ab326 heavy chain; Spike glycoprotein; Ab326 light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7x91 | 4.30 Å | 2022-12-07 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | phenix.real_space_refine; PHENIX | 2022-03-15 | 36406861 | - | - | - | - | N/A | - | - | Spike glycoprotein; An Fv-clasp version of the Ab496 light chain; An Fv-clasp version of the Ab496 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7x92 | 4.10 Å | 2022-12-07 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | phenix.real_space_refine; PHENIX | 2022-03-15 | 36406861 | - | - | - | - | N/A | - | - | Spike glycoprotein; Ab445 light chain; Ab445 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7zv7 | 1.34 Å | 2022-12-07 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 20.11% | 22.00% | 2.80% | inhibitor 57 | - | autoPROC | 94.20% | 18.0 | ESRF (MASSIF-1) | BUSTER | 2022-05-13 | 36332548 | - | - | - | - | N/A | - | - | inhibitor 57; 3C-like proteinase nsp5 | 64.4 | - | No | - | 65.0 | 42.1 | 96.7 | - | |||||||
| 7zv8 | 1.94 Å | 2022-12-07 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 19.18% | 23.08% | 5.20% | inhibitor 58 | - | autoPROC | 94.00% | 10.0 | ESRF (MASSIF-1) | BUSTER | 2022-05-13 | 36332548 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5; inhibitor 58 | 78.2 | - | No | - | 54.3 | 91.0 | 91.6 | - | |||||||
| 8bsd | 1.95 Å | 2022-12-07 | X-ray | TBN, MES | SARS-CoV-2 | NSP10/NSP16 | 18.04% | 20.75% | 6.39% | '2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | '2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | XDS | 98.86% | 8.3 | PETRA III, DESY (P11) | PHENIX; PHENIX | 2022-11-24 | - | ZN | - | - | - | N/A | - | - | 2'-O-methyltransferase nsp16; Non-structural protein 10 | 74.2 | - | No | - | - | 76.2 | 73.1 | 76.6 | - | ||||||
| 8dw2 | 4.11 Å | 2022-12-07 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | SR22 | - | - | - | () | PHENIX | 2022-07-30 | 36411391 | - | - | - | - | N/A | - | - | Antibody S309 light chain; SR22; Spike protein S1 - receptor binding domain (UNP residues 330-526); Antibody CR3022 light chain; Antibody CR3022 heavy chain; Antibody S309 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dw3 | 4.26 Å | 2022-12-07 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-07-30 | 36411391 | - | - | - | - | N/A | - | - | antibody CR3022 heavy chain; antibody S309 heavy chain; Spike protein S1 - receptor binding domain (UNP residues 330-526); Anti-SARS-CoV-2 DARPin SR16m; Antibody S309 light chain; antibody CR3022 light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8dw9 | 4.00 Å | 2022-12-07 | X-ray | - | SARS-CoV-2 | Spike | 30.00% | 38.07% | 95.70% | - | XDS | 99.80% | 2.9 | APS (24-ID-C) | PHENIX | 2022-08-01 | 36354024 | - | - | - | - | N/A | - | - | D29 Heavy chain; D29 Fab light chain; Spike protein S1 - receptor binding domain (UNP residues 333-527) | - | - | No | - | 3.5 | - | 68.0 | - | ||||||||
| 8dwa | 3.20 Å | 2022-12-07 | X-ray | NAG | SARS-CoV-2 | Spike | 23.48% | 29.75% | 51.60% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 100.00% | 6.9 | APS (24-ID-C) | REFMAC | 2022-08-01 | 36354024 | - | - | - | - | N/A | - | - | P1D9 Heavy chain; P1D9 Light chain; Spike protein S1 - receptor binding domain (UNP residues 335-515) | 4.4 | - | No | - | 7.9 | 46.3 | 13.2 | - | |||||||
| 8dxs | 3.76 Å | 2022-12-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-08-03 | 36354024 | - | - | - | - | N/A | - | - | P2B4 Heavy chain; P2B4 Light chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8ey2 | 3.50 Å | 2022-12-07 | Cryo-EM | - | SARS-CoV-2 | NSP5 (3CLpro) | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-10-26 | - | - | - | - | - | N/A | - | - | 3C-like proteinase - UNP residues 3259-3569 | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 8gwb | 2.75 Å | 2022-12-07 | Cryo-EM | MN | SARS-CoV-2 | NSP7/NSP8/NSP9/NSP12/NSP13 | N/A | N/A | N/A | MANGANESE (II) ION | MANGANESE (II) ION | - | - | () | PHENIX | 2022-09-16 | 36335936 | ZN; MN | - | - | - | N/A | - | - | Non-structural protein 8 - UNP residues 3943-4140; RNA-directed RNA polymerase - UNP residues 4393-5324; Non-structural protein 7 - UNP residues 3860-3942; primer; template; Helicase - UNP residues 5325-5925; Non-structural protein 9 - UNP residues 4141-4253; RNA (5'-R(P*AP*U)-3') | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8gz5 | 1.70 Å | 2022-12-07 | X-ray | - | SARS-CoV-2 | Spike | 17.13% | 18.90% | N/A | - | XDS | 100.00% | 12.2 | SPring-8 (BL44B2) | PHENIX | 2022-09-25 | 36413757 | - | - | - | - | N/A | - | - | Nanobody P17; Spike protein S1 | 85.3 | - | No | - | 88.2 | 93.5 | 85.9 | - | ||||||||
| 7w9g | 2.50 Å | 2022-12-14 | X-ray | SE | SARS-CoV-2 | NSP5 (3CLpro) | 19.50% | 24.68% | N/A | SELENIUM ATOM | SELENIUM ATOM | xia2 | 100.00% | 8.9 | RRCAT INDUS-2 (PX-BL21) | PHENIX; PHENIX | 2021-12-09 | 36650888 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 63.3 | - | No | - | 38.5 | 78.2 | 84.7 | - | |||||||
| 7wtf | 3.00 Å | 2022-12-14 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-04 | 35672388 | - | - | - | - | N/A | - | - | Spike glycoprotein; Light chain of XGv051; Heavy chain of XGv051 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wtg | 3.80 Å | 2022-12-14 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-02-04 | 35672388 | - | - | - | - | N/A | - | - | Light chain of XGv051; Heavy chain of XGv051; Spike protein S1 - RBD | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7wth | 4.30 Å | 2022-12-14 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-02-04 | 35672388 | - | - | - | - | N/A | - | - | Heavy chain of XGv264; Light chain of XGv264; Spike protein S1 - RBD | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7wtj | 4.20 Å | 2022-12-14 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-02-04 | 35672388 | - | - | - | - | N/A | - | - | Heavy chain of XGv286; Spike protein S1 - RBD; Light chain of XGv286 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7wtk | 3.60 Å | 2022-12-14 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-04 | 35672388 | - | - | - | - | N/A | - | - | Heavy chain of XGv286; Spike glycoprotein; Light chain of XGv286 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7x2m | 1.80 Å | 2022-12-14 | X-ray | - | SARS-CoV-2 | Spike | 17.96% | 20.72% | N/A | - | HKL-3000 | 99.93% | 33.27 | SSRF (BL18U1) | PHENIX | 2022-02-25 | 36575191 | - | - | - | - | N/A | - | - | Spike protein S1 - RBD; 1-2C7 | 43.5 | - | No | - | 76.3 | 14.9 | 71.3 | - | ||||||||
| 8gwg | 3.37 Å | 2022-12-14 | Cryo-EM | GTP | SARS-CoV-2 | NSP7/NSP8/NSP9/NSP12/NSP13 | N/A | N/A | N/A | Template; primer; GUANOSINE-5'-TRIPHOSPHATE | GUANOSINE-5'-TRIPHOSPHATE | - | - | () | PHENIX | 2022-09-17 | 36335936 | ZN | - | - | - | N/A | - | - | RNA-directed RNA polymerase - UNP residues 4393-5324; Non-structural protein 8 - UNP residues 3943-4140; Non-structural protein 7 - UNP residues 3860-3942; primer; Template; Helicase - UNP residues 5325-5925; Non-structural protein 9 - UNP residues 4141-4253 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8gwi | 3.18 Å | 2022-12-14 | Cryo-EM | GTP | SARS-CoV-2 | NSP7/NSP8/NSP9/NSP12/NSP13 | N/A | N/A | N/A | RNA (27-MER); GUANOSINE-5'-TRIPHOSPHATE; primer | GUANOSINE-5'-TRIPHOSPHATE | - | - | () | PHENIX | 2022-09-17 | 36335936 | ZN | - | - | - | N/A | - | - | Non-structural protein 8 - UNP residues 3943-4140; Non-structural protein 9; Helicase - UNP residues 5325-5925; RNA (27-MER); primer; Non-structural protein 7 - UNP residues 3860-3942; RNA-directed RNA polymerase - UNP residues 4393-5324 | N/A | - | No | - | N/A | N/A | N/A | RNA (27-MER) | - | |||||||
| 8gwn | 3.38 Å | 2022-12-14 | Cryo-EM | GNP | SARS-CoV-2 | NSP7/NSP8/NSP9/NSP12/NSP13 | N/A | N/A | N/A | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | - | - | () | PHENIX | 2022-09-17 | 36335936 | ZN | - | - | - | N/A | - | - | Non-structural protein 9; RNA-directed RNA polymerase - UNP residues 4393-5324; Non-structural protein 8 - UNP residues 3943-4140; Non-structural protein 7 - UNP residues 3860-3942; primer; template; Helicase - UNP residues 5325-5925 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7t72 | 3.18 Å | 2022-12-21 | X-ray | NAG | SARS-CoV-2 | Spike | 23.71% | 30.84% | 7.90% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.30% | 21.4 | Australian Synchrotron (MX2) | PHENIX | 2021-12-14 | 36755042 | - | - | - | - | N/A | - | - | Antibody light chain; Spike protein S1 - Receptor Binding Domain (RBD); Antibody heavy chain | 26.5 | - | No | - | 6.1 | 81.4 | 41.0 | - | |||||||
| 7wd1 | 2.50 Å | 2022-12-21 | X-ray | - | SARS-CoV-2 | Spike | 20.06% | 23.95% | 18.00% | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | - | HKL-2000 | 100.00% | 14.229 | SSRF (BL17U) | PHENIX | 2021-12-20 | 36702124 | - | - | - | - | N/A | - | - | Spike protein S1; R14 | 45.3 | - | No | - | 45.4 | 25.2 | 95.5 | - | |||||||
| 7wd2 | 2.69 Å | 2022-12-21 | X-ray | NAG | SARS-CoV-2 | Spike | 20.38% | 22.93% | 17.60% | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 100.00% | 16.4 | SSRF (BL17U) | PHENIX | 2021-12-20 | 36702124 | - | - | - | - | N/A | - | - | Anti-RON nanobody; Spike protein S1 | 50.7 | - | No | - | 55.7 | 36.4 | 84.3 | - | |||||||
| 7wnm | 2.70 Å | 2022-12-21 | X-ray | NAG | SARS-CoV-2 | Spike | 19.38% | 22.25% | 17.50% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.70% | 12.7 | SSRF (BL02U1) | PHENIX | 2022-01-18 | 35013100 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike protein S1 - Receptor-binding domain | 42.7 | - | No | - | 62.7 | 38.2 | 60.2 | - | |||||||
| 7wti | 3.80 Å | 2022-12-21 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-04 | 35672388 | - | - | - | - | N/A | - | - | Heavy chain of XGv264; Light chain of XGv264; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7x2j | 2.40 Å | 2022-12-21 | X-ray | NAG | SARS-CoV | Spike | 19.69% | 23.48% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-3000 | 99.77% | 25.45 | SSRF (BL18U1) | PHENIX | 2022-02-25 | 36575191 | - | - | - | - | N/A | - | - | Spike protein S1; Nb70 | 27.3 | - | No | - | 50.3 | 50.8 | 29.1 | - | |||||||
| 7x2k | 2.40 Å | 2022-12-21 | X-ray | NAG | SARS-CoV-2 | Spike | 19.50% | 24.49% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-3000 | 99.54% | 17.18 | SSRF (BL18U1) | PHENIX | 2022-02-25 | 36575191 | - | - | - | - | N/A | - | - | 1F11-L; Spike protein S1 - RBD; 1F11-H; Nb70 | 38.9 | - | No | - | 40.4 | 76.2 | 37.1 | - | ||||||||
| 7xa7 | 3.31 Å | 2022-12-21 | X-ray | NAG | SARS-CoV-2 | Spike | 23.28% | 25.37% | 21.80% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 96.50% | 8.8 | NFPSS (BL19U1) | PHENIX | 2022-03-17 | 35874946 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme; Spike protein S1 - Receptor binding domain | 28.1 | - | No | - | 32.0 | 41.6 | 58.3 | - | |||||||
| 7xrs | 1.93 Å | 2022-12-21 | X-ray | HUR | SARS-CoV-2 | NSP5 (3CLpro) | 19.96% | 24.31% | 5.70% | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | autoPROC | 98.90% | 8.1 | SSRF (BL02U1) | PHENIX | 2022-05-11 | 36146880 | - | - | - | - | N/A | - | - | Replicase polyprotein 1a | 66.2 | - | No | - | 41.9 | 93.1 | 72.4 | - | |||||||
| 7xry | 1.99 Å | 2022-12-21 | X-ray | HUR | MERS-CoV | NSP5 (3CLpro) | 22.36% | 25.35% | 6.20% | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | autoPROC | 99.90% | 11.0 | SSRF (BL02U1) | PHENIX | 2022-05-12 | 36146880 | - | - | - | - | N/A | - | - | ORF1a | 17.2 | - | No | - | 32.2 | 16.1 | 59.8 | - | |||||||
| 7ygq | 2.04 Å | 2022-12-21 | X-ray | HUR | SARS-CoV | NSP5 (3CLpro) | 20.54% | 23.50% | 2.80% | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | autoPROC | 96.40% | 9.3 | SSRF (BL02U1) | PHENIX; PHENIX | 2022-07-12 | 36146880 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 42.2 | - | No | - | 50.2 | 59.3 | 50.8 | - | |||||||
| 8d47 | 2.00 Å | 2022-12-21 | X-ray | PO4 | SARS-CoV-2 | Spike | 18.36% | 23.78% | 11.70% | SARS-CoV-2 fusion peptide; PHOSPHATE ION | PHOSPHATE ION | XDS | 97.90% | 6.1 | SSRL (BL12-1) | PHENIX | 2022-06-01 | 36701425 | - | - | - | - | N/A | - | - | fp.006 heavy chain; fp.006 light chain; SARS-CoV-2 fusion peptide | 37.9 | - | No | - | 47.2 | 53.3 | 51.4 | - | |||||||
| 8hda | 1.93 Å | 2022-12-21 | X-ray | - | SARS-CoV-2 | NSP3: Ubl1 | 18.41% | 22.87% | 9.40% | - | HKL-3000 | 99.30% | 10.7 | SSRF (BL19U1) | REFMAC | 2022-11-03 | - | - | - | - | - | N/A | - | - | Papain-like protease nsp3 | 62.8 | - | No | - | - | 56.4 | 59.4 | 84.4 | - | |||||||
| 7qka | 1.80 Å | 2022-12-28 | X-ray | UED | SARS-CoV-2 | NSP5 (3CLpro) | 16.30% | 20.72% | 4.32% | N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | XDS | 98.89% | 23.4 | PETRA III, DESY (P11) | PHENIX | 2021-12-17 | 37853179 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 73.4 | - | No | - | 76.3 | 73.6 | 73.8 | - | |||||||
| 7vhh | 3.80 Å | 2022-12-28 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-09-22 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7wch | 3.19 Å | 2022-12-28 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2021-12-20 | - | - | - | - | - | N/A | - | - | SWA9H; SWA9L; Surface glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||||
| 7wck | 3.49 Å | 2022-12-28 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-12-20 | - | - | - | - | - | N/A | - | - | SWA9H; Surface glycoprotein; SWA9L | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7wcp | 3.01 Å | 2022-12-28 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-12-20 | - | - | - | - | - | N/A | - | - | SWC11L; Surface glycoprotein; SWC11H | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7wcu | 3.09 Å | 2022-12-28 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-12-20 | - | - | - | - | - | N/A | - | - | Surface glycoprotein; SWC11H; SWC11L | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7wfc | 2.60 Å | 2022-12-28 | X-ray | - | HKU1-CoV | NSP3: PLpro | 17.11% | 25.11% | 9.20% | - | CrysalisPro | 99.31% | 11.77 | ROTATING ANODE () | PHENIX | 2021-12-26 | 37130166 | ZN | - | - | - | N/A | - | - | 60S ribosomal protein L40; Papain-like protease | 31.4 | - | No | - | 34.1 | 46.4 | 58.3 | - | ||||||||
| 7wo2 | 1.96 Å | 2022-12-28 | X-ray | 40I | SARS-CoV-2 | NSP5 (3CLpro) | 18.20% | 21.75% | N/A | N-[(2S)-3-methyl-1-[[(2S)-4-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanylidene-3-[(3S}-2-oxidanylidenepiperidin-3-yl]propan-2-yl]amino]pentan-2-yl]amino]-1-oxidanylidene-butan-2-yl]furan-2-carboxamide | N-[(2S)-3-methyl-1-[[(2S)-4-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanylidene-3-[(3S}-2-oxidanylidenepiperidin-3-yl]propan-2-yl]amino]pentan-2-yl]amino]-1-oxidanylidene-butan-2-yl]furan-2-carboxamide | 100.00% | 53.7 | SSRF (BL19U1) | PHENIX | 2022-01-20 | 35216507 | - | - | - | - | N/A | - | - | 3C-like proteinase | 42.6 | - | No | - | 67.5 | 32.6 | 60.8 | - | ||||||||
| 7ykj | 3.50 Å | 2022-12-28 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-07-22 | 36354024 | - | - | - | - | N/A | - | - | P3E6 heavy chain; Spike glycoprotein; P3E6 light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7zqv | 2.26 Å | 2022-12-28 | X-ray | XNV | SARS-CoV-2 | NSP5 (3CLpro) | 19.02% | 24.05% | N/A | ethyl (4R)-4-({(2S)-2-[3-{[(5-methyl-1,2-oxazol-3-yl)carbonyl]amino}-2-oxopyridin-1(2H)-yl]pent-4-ynoyl}amino)-5-[(3S)-2-oxopyrrolidin-3-yl]pentanoate | XDS | 89.50% | 4.5 | ESRF (BM30A) | REFMAC | 2022-05-03 | 36336176 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 31.0 | - | No | - | 44.4 | 54.1 | 39.4 | - | ||||||||
| 7zqw | 2.53 Å | 2022-12-28 | X-ray | XNV | SARS-CoV | NSP5 (3CLpro) | 21.54% | 28.91% | N/A | ethyl (4R)-4-({(2S)-2-[3-{[(5-methyl-1,2-oxazol-3-yl)carbonyl]amino}-2-oxopyridin-1(2H)-yl]pent-4-ynoyl}amino)-5-[(3S)-2-oxopyrrolidin-3-yl]pentanoate | ethyl (4R)-4-({(2S)-2-[3-{[(5-methyl-1,2-oxazol-3-yl)carbonyl]amino}-2-oxopyridin-1(2H)-yl]pent-4-ynoyl}amino)-5-[(3S)-2-oxopyrrolidin-3-yl]pentanoate | XDS | 91.90% | 10.1 | ALBA (XALOC) | REFMAC | 2022-05-03 | 36336176 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 12.0 | - | No | - | 10.2 | 59.6 | 24.7 | - | |||||||
| 8aou | - | 2022-12-28 | NMR | - | SARS-CoV-2 | NSP1 | N/A | N/A | N/A | - | - | - | () | 2022-08-08 | 36610391 | - | - | - | - | N/A | - | - | Host translation inhibitor nsp1 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 8b0s | 2.42 Å | 2022-12-28 | X-ray | AU | SARS-CoV-2 | NSP5 (3CLpro) | 25.35% | 27.27% | 16.00% | GOLD ION | GOLD ION | XDS | 94.50% | 5.3 | SEALED TUBE () | PHENIX | 2022-09-08 | 36421689 | AU | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 8.0 | - | No | - | 18.1 | 30.8 | 32.9 | - | |||||||
| 8b0t | 2.40 Å | 2022-12-28 | X-ray | AU | SARS-CoV-2 | NSP5 (3CLpro) | 23.73% | 27.55% | 18.00% | GOLD ION | GOLD ION | XDS | 94.10% | 5.9 | SEALED TUBE () | PHENIX | 2022-09-08 | 36421689 | AU | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | 8.1 | - | No | - | 16.4 | 23.7 | 42.1 | - | |||||||
| 7wgv | 3.20 Å | 2023-01-04 | Cryo-EM | BLR, EIC, NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid; LINOLEIC ACID | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid; LINOLEIC ACID; 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-12-29 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7wgx | 3.50 Å | 2023-01-04 | Cryo-EM | BLR, NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid; 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-12-29 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7wgy | 4.00 Å | 2023-01-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-12-29 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7wgz | 4.50 Å | 2023-01-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2021-12-29 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7y76 | 3.20 Å | 2023-01-04 | Cryo-EM | 3PH, NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose; 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE; 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-06-21 | 36384914 | ZN | - | - | - | N/A | - | - | SIT1; Angiotensin-converting enzyme 2; Spike protein S1 - BA.4 RBD | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7tek | 2.20 Å | 2023-01-11 | X-ray | I2D | SARS-CoV-2 | NSP5 (3CLpro) | 20.80% | 27.38% | N/A | N-[(3-chlorophenyl)methyl]-N-[4-(1H-pyrazol-4-yl)phenyl]-2-(pyridin-3-yl)acetamide | N-[(3-chlorophenyl)methyl]-N-[4-(1H-pyrazol-4-yl)phenyl]-2-(pyridin-3-yl)acetamide | HKL-2000 | 99.37% | 26.62 | APS (21-ID-F) | PHENIX | 2022-01-05 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 17.4 | - | - | - | ||||||
| 7tel | 2.40 Å | 2023-01-11 | X-ray | I2N | SARS-CoV-2 | NSP5 (3CLpro) | 19.89% | 25.99% | N/A | N-[(3-chloro-5-fluorophenyl)methyl]-N-[4-(1H-imidazol-4-yl)phenyl]-2-(isoquinolin-4-yl)acetamide | N-[(3-chloro-5-fluorophenyl)methyl]-N-[4-(1H-imidazol-4-yl)phenyl]-2-(isoquinolin-4-yl)acetamide | HKL-2000 | 98.48% | 15.85 | APS (21-ID-F) | PHENIX | 2022-01-05 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 26.8 | - | - | - | ||||||
| 7uzc | 2.20 Å | 2023-01-11 | X-ray | - | SARS-CoV-2 | Spike | 17.15% | 24.33% | 12.00% | - | XDS | 98.60% | 9.9 | SSRL (BL12-2) | PHENIX | 2022-05-08 | 36370711 | - | - | - | - | N/A | - | - | Spike protein S1 - RBD; M8a-34 Fab light chain; M8a-34 Fab heavy chain | - | - | No | - | 41.7 | - | - | - | ||||||||
| 7uzd | 3.00 Å | 2023-01-11 | X-ray | - | SARS-CoV-2 | Spike | 23.09% | 27.27% | 23.30% | - | XDS | 99.80% | 12.3 | SSRL (BL12-2) | PHENIX | 2022-05-08 | 36370711 | - | - | - | - | N/A | - | - | HSW-2 Fab light chain; HSW-2 Fab heavy chain; Spike protein S1 - RBD | - | - | No | - | 18.1 | - | - | - | ||||||||
| 7x4i | 3.38 Å | 2023-01-11 | X-ray | - | SARS-CoV | Spike | 26.70% | 32.09% | 20.00% | - | autoPROC | 99.50% | 8.7 | SSRF (BL02U1) | PHENIX | 2022-03-02 | 36494344 | - | - | - | - | N/A | - | - | Spike glycoprotein; nanobody aSA3 | - | - | No | - | 4.9 | - | - | - | ||||||||
| 7xbf | 3.51 Å | 2023-01-11 | X-ray | NAG | RshSTT182/200 | Spike | 19.93% | 23.27% | 13.30% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.90% | 9.0 | SSRF (BL02U1) | PHENIX | 2022-03-21 | 36519268 | ZN | - | - | - | N/A | - | - | RshSTT182/200 coronavirus receptor binding domain insert2; Processed angiotensin-converting enzyme 2 | - | - | No | - | 52.4 | - | - | - | |||||||
| 7xbg | 3.37 Å | 2023-01-11 | X-ray | NAG | RshSTT182/200 | Spike | 26.92% | 27.36% | 15.10% | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 100.00% | 10.5 | SSRF (BL02U1) | PHENIX | 2022-03-21 | 36519268 | ZN | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; RshSTT182/200 coronavirus receptor binding domain insert2 mutant | - | - | No | - | 17.5 | - | - | - | |||||||
| 7xbh | 3.02 Å | 2023-01-11 | X-ray | NAG | RshSTT182/200 | Spike | 20.06% | 23.46% | 16.60% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.80% | 9.4 | SSRF (BL02U1) | PHENIX | 2022-03-21 | 36519268 | ZN | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; RshSTT182/200 coronavirus receptor binding domain | - | - | No | - | 50.4 | - | - | - | |||||||
| 7xnf | 2.79 Å | 2023-01-11 | X-ray | - | Pangolin CoV | Spike | 31.96% | 37.95% | 15.30% | - | HKL-2000 | 99.80% | 11.905 | SSRF (BL19U1) | PHENIX | 2022-04-28 | 36493785 | - | - | - | - | N/A | - | - | P2C-1F11 Heavy Chain; P2C-1F11 Lambda chain; Spike protein S1 - BetaCoV S1-CTD | - | - | No | - | 3.5 | - | - | - | ||||||||
| 7xsw | 3.30 Å | 2023-01-11 | X-ray | - | Pangolin CoV | Spike | 27.66% | 32.16% | N/A | - | HKL-2000 | 89.00% | 1.9 | SSRF (BL19U1) | PHENIX | 2022-05-15 | 36493785 | - | - | - | - | N/A | - | - | S309 Heavy Chain; Spike protein S1 - BetaCoV S1-CTD; S309 Lambda Chain | - | - | No | - | 4.9 | - | - | - | ||||||||
| 8asy | 2.85 Å | 2023-01-11 | X-ray | PGE, NAG | SARS-CoV-2 | Spike | 21.73% | 26.47% | 44.30% | 2-acetamido-2-deoxy-beta-D-glucopyranose; TRIETHYLENE GLYCOL | TRIETHYLENE GLYCOL; 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 99.80% | 7.6 | Diamond (I03) | PHENIX | 2022-08-22 | 36586406 | - | - | - | - | N/A | - | - | Spike protein S1; Processed angiotensin-converting enzyme 2 | - | - | No | - | 23.3 | - | - | - | |||||||
| 8fd5 | 4.57 Å | 2023-01-11 | Cryo-EM | - | SARS-CoV-2 | Nucleocapsid | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-12-02 | 37749713 | - | - | - | - | N/A | - | - | Nucleoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8fg2 | 6.00 Å | 2023-01-11 | Cryo-EM | - | SARS-CoV-2 | Nucleocapsid | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-12-12 | 37749713 | - | - | - | - | N/A | - | - | Nucleoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8gwe | 2.66 Å | 2023-01-11 | Cryo-EM | GNP | SARS-CoV-2 | NSP7/NSP8/NSP9/NSP12/NSP13 | N/A | N/A | N/A | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | - | - | () | PHENIX | 2022-09-16 | 36335936 | MG; ZN | - | - | - | N/A | - | - | RNA-directed RNA polymerase - UNP residues 4393-5324; Non-structural protein 8 - UNP residues 3943-4140; Non-structural protein 7 - UNP residues 3860-3942; primer; template; Helicase - UNP residues 5325-5925; Non-structural protein 9 - UNP residues 4141-4253; RNA (5'-R(P*AP*UP*UP*A)-3') | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8gwf | 3.39 Å | 2023-01-11 | Cryo-EM | GTP | SARS-CoV-2 | NSP7/NSP8/NSP9/NSP12/NSP13 | N/A | N/A | N/A | GUANOSINE-5'-TRIPHOSPHATE; primer; template | GUANOSINE-5'-TRIPHOSPHATE | - | - | () | PHENIX | 2022-09-17 | 36335936 | ZN | - | - | - | N/A | - | - | RNA-directed RNA polymerase - UNP residues 4393-5324; Non-structural protein 8 - UNP residues 3943-4140; Non-structural protein 7 - UNP residues 3860-3942; primer; template; Helicase - UNP residues 5325-5925; Non-structural protein 9 - UNP residues 4141-4253 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7pxz | 1.75 Å | 2023-01-18 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 17.38% | 20.29% | N/A | - | CrystFEL | 100.00% | 11.443052 | FREE ELECTRON LASER () | PHENIX; REFMAC | 2021-10-08 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 79.8 | - | - | - | |||||||
| 7wh8 | 3.11 Å | 2023-01-18 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-12-30 | 37542391 | - | - | - | - | N/A | - | - | antibody ZB8 heavy chain; Spike glycoprotein; antibody ZB8 light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7whb | 2.67 Å | 2023-01-18 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-12-30 | 37542391 | - | - | - | - | N/A | - | - | Spike glycoprotein; antibody ZB8 light chain; antibody ZB8 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7whd | 2.65 Å | 2023-01-18 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2021-12-30 | 37542391 | - | - | - | - | N/A | - | - | Spike glycoprotein; antibody ZB8 heavy chain; antibody ZB8 light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7x25 | 2.49 Å | 2023-01-18 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2022-02-25 | 36246239 | - | - | - | - | N/A | - | - | antibody S41 heavy chain; Spike glycoprotein; antibody S41 light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 7x28 | 2.49 Å | 2023-01-18 | Cryo-EM | - | MERS-CoV | Spike | N/A | N/A | N/A | - | - | - | () | 2022-02-25 | 36246239 | - | - | - | - | N/A | - | - | antibody S41 light chain; antibody S41 heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 8bzv | 1.80 Å | 2023-01-18 | X-ray | ADN, MES | SARS-CoV-2 | NSP10/NSP16 | 17.76% | 19.95% | 13.20% | ADENOSINE; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | ADENOSINE; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | XDS | 99.90% | 6.64 | PETRA III, DESY (P11) | PHENIX; PHENIX | 2022-12-15 | - | ZN | - | - | - | N/A | - | - | 2'-O-methyltransferase nsp16; Non-structural protein 10 | - | - | No | - | - | 82.0 | - | - | - | ||||||
| 8dxt | 2.25 Å | 2023-01-18 | X-ray | - | SARS-CoV-2 | Spike | 21.12% | 25.36% | 6.00% | - | XDS | 100.00% | 17.9 | Australian Synchrotron (MX2) | PHENIX | 2022-08-03 | 36755042 | - | - | - | - | N/A | - | - | Spike protein S1 - receptor binding domain; Heavy chain of Fab arm of antibody GAR12; Light chain of Fab arm of antibody GAR12 | - | - | No | - | 32.1 | - | - | - | ||||||||
| 8dxu | 2.73 Å | 2023-01-18 | X-ray | NAG | SARS-CoV-2 | Spike | 22.29% | 26.90% | 15.60% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.40% | 12.8 | Australian Synchrotron (MX2) | PHENIX | 2022-08-03 | 36755042 | - | - | - | - | N/A | - | - | Heavy chain of Fab arm of antibody 10G4; Light chain of Fab arm of antibody 10G4; Heavy chain of Fab arm of antibody GAR03; Light chain of Fab arm of antibody GAR03; Spike protein S1 - receptor binding domain | - | - | No | - | 20.1 | - | - | - | |||||||
| 7mx9 | 2.60 Å | 2023-01-25 | X-ray | - | SARS-CoV-2 | ORF8 | 24.32% | 29.66% | 16.60% | - | XDS | 100.00% | 11.8 | ROTATING ANODE () | PHENIX | 2021-05-18 | - | - | - | - | - | N/A | - | - | ORF8 protein | - | - | No | - | - | 8.1 | - | - | - | |||||||
| 7pzq | 2.25 Å | 2023-01-25 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 17.74% | 23.89% | N/A | - | CrystFEL | 99.99% | 7.41 | FREE ELECTRON LASER () | PHENIX; PHENIX | 2021-10-13 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 46.1 | - | - | - | |||||||
| 7t5o | 3.39 Å | 2023-01-25 | Cryo-EM | - | SARS-CoV | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2021-12-12 | 36755042 | - | - | - | - | N/A | - | - | GAR03 Fab light chain; Spike glycoprotein; GAR03 Fab heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7uu6 | 1.85 Å | 2023-01-25 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 22.40% | 25.21% | N/A | - | PROTEUM PLUS | 99.90% | 14.1 | ROTATING ANODE () | PHENIX | 2022-04-28 | 36629751 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 33.2 | - | - | - | ||||||||
| 7uu7 | 2.49 Å | 2023-01-25 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 19.46% | 24.50% | N/A | - | PROTEUM PLUS | 98.20% | 19.4 | ROTATING ANODE () | PHENIX | 2022-04-28 | 36629751 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 40.0 | - | - | - | ||||||||
| 7uu8 | 2.50 Å | 2023-01-25 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 20.26% | 25.74% | N/A | - | PROTEUM PLUS | 99.30% | 13.3 | ROTATING ANODE () | PHENIX | 2022-04-28 | 36629751 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 28.8 | - | - | - | ||||||||
| 7uu9 | 2.47 Å | 2023-01-25 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 20.10% | 25.33% | N/A | - | PROTEUM PLUS | 99.20% | 14.2 | ROTATING ANODE () | PHENIX | 2022-04-28 | 36629751 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 32.2 | - | - | - | ||||||||
| 7uua | 1.85 Å | 2023-01-25 | X-ray | NOL | SARS-CoV-2 | NSP5 (3CLpro) | 24.37% | 27.30% | N/A | N-[(BENZYLOXY)CARBONYL]-O-(TERT-BUTYL)-L-THREONYL-3-CYCLOHEXYL-N-[(1S)-2-HYDROXY-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}ETHYL]-L-ALANINAMIDE | N-[(BENZYLOXY)CARBONYL]-O-(TERT-BUTYL)-L-THREONYL-3-CYCLOHEXYL-N-[(1S)-2-HYDROXY-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}ETHYL]-L-ALANINAMIDE | PROTEUM PLUS | 99.90% | 13.8 | ROTATING ANODE () | PHENIX | 2022-04-28 | 36629751 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 17.8 | - | - | - | |||||||
| 7uub | 1.63 Å | 2023-01-25 | X-ray | 7VB | SARS-CoV-2 | NSP5 (3CLpro) | 21.11% | 22.97% | N/A | N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methylidene-L-norvalinamide | N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methylidene-L-norvalinamide | iMOSFLM | 95.80% | 12.5 | ROTATING ANODE () | PHENIX | 2022-04-28 | 36629751 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 55.4 | - | - | - | |||||||
| 7uuc | 1.60 Å | 2023-01-25 | X-ray | 81L | SARS-CoV-2 | NSP5 (3CLpro) | 20.82% | 22.92% | N/A | N-[(benzyloxy)carbonyl]-3-methyl-L-valyl-3-cyclopropyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | N-[(benzyloxy)carbonyl]-3-methyl-L-valyl-3-cyclopropyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | iMOSFLM | 99.70% | 11.3 | ROTATING ANODE () | PHENIX | 2022-04-28 | 36629751 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 55.8 | - | - | - | |||||||
| 7uud | 1.85 Å | 2023-01-25 | X-ray | I71 | SARS-CoV-2 | NSP5 (3CLpro) | 20.93% | 23.69% | N/A | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-(ethylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-(ethylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | PROTEUM PLUS | 99.90% | 19.4 | ROTATING ANODE () | PHENIX | 2022-04-28 | 36629751 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 48.1 | - | - | - | |||||||
| 7uue | 1.85 Å | 2023-01-25 | X-ray | I65 | SARS-CoV-2 | NSP5 (3CLpro) | 20.51% | 23.18% | N/A | benzyl [(2S,3R)-1-({(2S)-1-[2-(3-amino-3-oxopropyl)-2-propanoylhydrazinyl]-3-cyclohexyl-1-oxopropan-2-yl}amino)-3-tert-butoxy-1-oxobutan-2-yl]carbamate (non-preferred name) | benzyl [(2S,3R)-1-({(2S)-1-[2-(3-amino-3-oxopropyl)-2-propanoylhydrazinyl]-3-cyclohexyl-1-oxopropan-2-yl}amino)-3-tert-butoxy-1-oxobutan-2-yl]carbamate (non-preferred name) | PROTEUM PLUS | 97.40% | 16.6 | ROTATING ANODE () | PHENIX | 2022-04-28 | 36629751 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 53.4 | - | - | - | |||||||
| 7wqi | 1.93 Å | 2023-01-25 | X-ray | 80I | SARS-CoV | NSP5 (3CLpro) | 21.43% | 23.98% | 4.20% | [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | XDS | 96.40% | 8.9 | SSRF (BL02U1) | PHENIX | 2022-01-25 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 45.2 | - | - | - | ||||||
| 7wqv | 2.80 Å | 2023-01-25 | X-ray | NAG, CA, MPD | SARS-CoV-2 | Spike | 21.98% | 25.10% | 16.10% | alpha-L-fucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose; alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose; (4S)-2-METHYL-2,4-PENTANEDIOL; CALCIUM ION; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose; CALCIUM ION; (4S)-2-METHYL-2,4-PENTANEDIOL | XDS | 99.30% | 9.4 | SSRF (BL18U1) | PHENIX | 2022-01-26 | 36706160 | CA | - | - | - | N/A | - | - | Ab08; Spike protein S1 - RBD | - | - | No | - | 34.2 | - | - | - | |||||||
| 8be1 | 1.98 Å | 2023-01-25 | X-ray | - | SARS-CoV-2 | Spike | 24.75% | 28.40% | 10.90% | - | xia2 | 100.00% | 13.2 | Diamond (I04) | BUSTER | 2022-10-21 | - | - | - | - | - | N/A | - | - | Antibody heavy chain; Antibody light chain; Spike protein S1 | - | - | No | - | - | 12.3 | - | - | - | |||||||
| 8cx9 | 3.50 Å | 2023-01-25 | X-ray | BR, NA | SARS-CoV-2 | NSP3: PLpro | 22.49% | 26.97% | 12.40% | BROMIDE ION; SODIUM ION | BROMIDE ION; SODIUM ION | MOSFLM | 81.90% | 6.4 | ROTATING ANODE () | PHENIX | 2022-05-20 | 36548304 | NA; ZN | - | - | - | N/A | - | - | Papain-like protease nsp3; Ubiquitin variant UbV.CV2.1 | - | - | No | - | 19.8 | - | - | - | |||||||
| 8d48 | 3.70 Å | 2023-01-25 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-06-01 | 36701425 | - | - | - | - | N/A | - | - | Spike glycoprotein; sd1.040 Fab light chain; sd1.040 Fab heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8dgu | 1.89 Å | 2023-01-25 | X-ray | - | Spike | 21.44% | 24.51% | 9.10% | Spike protein S2' | - | HKL-2000 | 98.20% | 7.2 | APS (23-ID-D) | PHENIX | 2022-06-24 | 36889306 | - | - | - | - | N/A | - | - | Spike protein S2' - Stem helix peptide, residues 1140-1164; Antibody CC25.106 Fab light chain; Antibody CC25.106 Fab heavy chain | - | - | No | - | 39.9 | - | - | - | ||||||||
| 8dm1 | 3.04 Å | 2023-01-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-07-08 | 36640338 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dm2 | 2.91 Å | 2023-01-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-07-08 | 36640338 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dm3 | 2.37 Å | 2023-01-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-07-08 | 36640338 | - | - | - | - | N/A | - | - | Fab 4A8 light chain; Fab 4A8 heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dm4 | 2.45 Å | 2023-01-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-07-08 | 36640338 | - | - | - | - | N/A | - | - | Spike glycoprotein; Fab 4A8 light chain; Fab 4A8 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8hc2 | 6.21 Å | 2023-01-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-11-01 | 36828833 | - | - | - | - | N/A | - | - | Spike glycoprotein; Heavy chain of YB9-258 Fab; Light chain of YB9-258 Fab | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8hc3 | 4.35 Å | 2023-01-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-11-01 | 36828833 | - | - | - | - | N/A | - | - | Spike glycoprotein; Heavy chain of YB9-258; Light chain of YB9-258 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8hc4 | 3.54 Å | 2023-01-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-11-01 | 36828833 | - | - | - | - | N/A | - | - | Light chain of YB9-258 Fab; Light chain of R1-32 Fab; Heavy chain of YB9-258; Heavy chain of R1-32 Fab; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8hc5 | 3.43 Å | 2023-01-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-11-01 | 36828833 | - | - | - | - | N/A | - | - | Light chain of YB9-258; Heavy chain of YB9-258 Fab; Spike glycoprotein; Light chain of R1-32 Fab; Heavy chain of R1-32 Fab | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8hc6 | 4.69 Å | 2023-01-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-11-01 | 36828833 | - | - | - | - | N/A | - | - | Spike protein S1; Heavy chain of YB9-258 Fab; Spike protein S1; Light chain of YB9-258 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8hc7 | 4.54 Å | 2023-01-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-11-01 | 36828833 | - | - | - | - | N/A | - | - | Spike protein S1; Light chain variable region of YB9-258; Spike protein S1; Heavy chain variable region of YB9-258 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8hc8 | 3.95 Å | 2023-01-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-11-01 | 36828833 | - | - | - | - | N/A | - | - | Spike protein S1; Light chain of YB13-292 Fab; Heavy chain of YB13-292 Fab | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8hc9 | 6.03 Å | 2023-01-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-11-01 | 36828833 | - | - | - | - | N/A | - | - | Spike glycoprotein; Light chain of YB13-292 Fab; Heavy chain of YB13-292 Fab | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8hca | 4.35 Å | 2023-01-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-11-01 | 36828833 | - | - | - | - | N/A | - | - | Light chain of YB13-292 Fab; Spike glycoprotein; Heavy chain of YB13-292 Fab | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8hcb | 4.18 Å | 2023-01-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-11-01 | 36828833 | - | - | - | - | N/A | - | - | Light chain of YB13-292 Fab; Spike glycoprotein; Heavy chain of YB13-292 Fab | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wr9 | 3.24 Å | 2023-02-01 | Cryo-EM | NAG | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-26 | - | - | - | - | - | N/A | - | - | Spike protein S1; BD55-3152H; BD55-3152L | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7wrh | 2.66 Å | 2023-02-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-26 | 35821014 | ZN | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wzo | 2.64 Å | 2023-02-01 | X-ray | - | SARS-CoV-2 | NSP3(Ubl1)/Nucleocapsid | 21.43% | 24.68% | 15.00% | - | XDS | 99.70% | 13.2 | SSRF (BL19U1) | PHENIX | 2022-02-18 | 36806252 | - | - | - | - | N/A | - | - | Nucleoprotein; nsp3 - ubiquitin-like domain 1 | - | - | No | - | 38.5 | - | - | - | ||||||||
| 7yck | 2.60 Å | 2023-02-01 | X-ray | - | SARS-CoV-2 | Spike | 22.85% | 26.44% | 9.30% | - | MOSFLM | 97.80% | 5.6 | NSRRC (BL15A1) | PHENIX | 2022-07-01 | 36658148 | - | - | - | - | N/A | - | - | FP-12A Fab light chain; Spike protein S1; FP-12A Fab heavy chain | - | - | No | - | 23.4 | - | - | - | ||||||||
| 7ycl | 2.13 Å | 2023-02-01 | X-ray | - | SARS-CoV-2 | Spike | 17.35% | 21.75% | 8.40% | - | MOSFLM | 99.70% | 9.7 | NSRRC (BL15A1) | PHENIX | 2022-07-01 | 36658148 | - | - | - | - | N/A | - | - | IS-9A Fab light chain; Spike protein S1; IS-9A Fab heavy chain | - | - | No | - | 67.5 | - | - | - | ||||||||
| 7ycn | 2.85 Å | 2023-02-01 | X-ray | NAG | SARS-CoV-2 | Spike | 21.14% | 24.99% | 15.10% | 2-acetamido-2-deoxy-beta-D-glucopyranose | MOSFLM | 99.90% | 6.6 | NSRRC (BL15A1) | PHENIX | 2022-07-01 | 36658148 | - | - | - | - | N/A | - | - | IY-2A Fab heavy chain; Spike protein S1; IY-2A Fab light chain | - | - | No | - | 35.4 | - | - | - | ||||||||
| 8hhx | 3.62 Å | 2023-02-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-11-17 | 36658148 | - | - | - | - | N/A | - | - | Spike glycoprotein; FP-12A Fab heavy chain; FP-12A Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8hhy | 2.77 Å | 2023-02-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-11-17 | 36658148 | - | - | - | - | N/A | - | - | IS-9A Fab heavy chain; IS-9A Fab light chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8hhz | 4.28 Å | 2023-02-01 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2022-11-17 | 36658148 | - | - | - | - | N/A | - | - | IY-2A Fab heavy chain; IY-2A Fab light chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 7tpk | 3.40 Å | 2023-02-08 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-01-25 | - | - | - | - | - | N/A | - | - | Spike protein S1 - Receptor-binding domain | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7wul | 1.58 Å | 2023-02-08 | X-ray | - | 15.97% | 19.28% | 5.80% | 3-mer peptide | - | HKL-2000 | 99.50% | 10.3 | NSRRC (TPS 05A) | PHENIX | 2022-02-08 | - | ZN | - | - | - | N/A | - | - | 3-mer peptide; E3 ubiquitin-protein ligase | - | - | No | - | - | 86.3 | - | - | - | ||||||||
| 7xwx | 3.00 Å | 2023-02-08 | X-ray | PO4 | SARS-CoV-2 | Nucleocapsid | 21.92% | 26.81% | 22.40% | PHOSPHATE ION | PHOSPHATE ION | XDS | 99.60% | 12.0 | SSRF (BL18U1) | REFMAC | 2022-05-27 | 36317101 | - | - | - | - | N/A | - | - | Nucleoprotein - C-terminal domain | - | - | No | - | 20.7 | - | - | - | |||||||
| 7xwz | 2.25 Å | 2023-02-08 | X-ray | - | SARS-CoV-2 | Nucleocapsid/RNA/RNA | 22.64% | 26.85% | 4.90% | RNA (5'-R(*CP*AP*CP*UP*GP*AP*C)-3'); RNA (5'-R(P*GP*UP*CP*AP*GP*UP*G)-3') | - | XDS | 99.60% | 17.7 | CLSI (08ID-1) | REFMAC | 2022-05-27 | 36317101 | - | - | - | - | N/A | - | - | RNA (5'-R(P*GP*UP*CP*AP*GP*UP*G)-3'); RNA (5'-R(*CP*AP*CP*UP*GP*AP*C)-3'); Nucleoprotein - N-terminal domain | - | - | No | - | 20.5 | - | - | RNA (5'-R(*CP*AP*CP*UP*GP*AP*C)-3'); RNA (5'-R(P*GP*UP*CP*AP*GP*UP*G)-3') | - | ||||||
| 7xx1 | 1.90 Å | 2023-02-08 | X-ray | MES | SARS-CoV-2 | Nucleocapsid | 20.96% | 25.84% | 6.40% | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | XDS | 99.60% | 10.6 | NSLS-II (17-ID-2) | REFMAC | 2022-05-27 | 36317101 | ZN | - | - | - | N/A | - | - | Nucleoprotein - N-terminal domain | - | - | No | - | 27.9 | - | - | - | |||||||
| 8dm5 | 2.51 Å | 2023-02-08 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-07-08 | 36640338 | - | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dm6 | 2.77 Å | 2023-02-08 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-07-08 | 36640338 | - | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dm7 | 2.49 Å | 2023-02-08 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-07-08 | 36640338 | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dm8 | 2.68 Å | 2023-02-08 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-07-08 | 36640338 | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dm9 | 2.56 Å | 2023-02-08 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-07-08 | 36640338 | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dma | 2.79 Å | 2023-02-08 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-07-08 | 36640338 | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8dqu | 2.45 Å | 2023-02-08 | X-ray | - | SARS-CoV-2 | NSP9 | 20.32% | 22.89% | N/A | - | XDS | 95.30% | 10.6 | Australian Synchrotron (MX2) | PHENIX | 2022-07-20 | 37036856 | - | - | - | - | N/A | - | - | Nanobody; Non-structural protein 9 | - | - | No | - | 56.3 | - | - | - | ||||||||
| 8eqj | 3.00 Å | 2023-02-08 | Cryo-EM | PEE | SARS-CoV-2 | ORF3a | N/A | N/A | N/A | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine | - | - | () | 2022-10-07 | 36695574 | - | - | - | - | N/A | - | - | ORF3a protein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8eqs | 3.10 Å | 2023-02-08 | Cryo-EM | PEE | ORF3a | N/A | N/A | N/A | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine | - | - | () | 2022-10-09 | 36695574 | - | - | - | - | N/A | - | - | ORF3a protein; Apolipoprotein A-I | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 8eqt | 3.40 Å | 2023-02-08 | Cryo-EM | PEE | SARS-CoV-2 | ORF3a | N/A | N/A | N/A | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine | - | - | () | 2022-10-09 | 36695574 | - | - | - | - | N/A | - | - | ORF3a protein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8equ | 2.80 Å | 2023-02-08 | Cryo-EM | PEE | SARS-CoV-2 | ORF3a | N/A | N/A | N/A | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine | - | - | () | PHENIX | 2022-10-09 | 36695574 | - | - | - | - | N/A | - | - | Saposin-A; Saposin A, polyalanine model; ORF3a protein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wwi | 3.50 Å | 2023-02-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-13 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; 55A8 heavy chain; 55A8 light chain | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7wwj | 3.50 Å | 2023-02-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-13 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; 55A8 light chain; 55A8 heavy chain | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7wwk | 3.40 Å | 2023-02-15 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-02-13 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; 55A8 light chain; 55A8 light chain | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7zh1 | 2.48 Å | 2023-02-15 | Cryo-EM | NAG, EIC | SARS-CoV | Spike | N/A | N/A | N/A | LINOLEIC ACID; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose; LINOLEIC ACID | - | - | () | PHENIX | 2022-04-05 | 36417532 | - | - | - | - | N/A | - | - | Spike glycoprotein,Fibritin | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7zh2 | 2.71 Å | 2023-02-15 | Cryo-EM | NAG, EIC | SARS-CoV | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; LINOLEIC ACID | 2-acetamido-2-deoxy-beta-D-glucopyranose; LINOLEIC ACID | - | - | () | PHENIX | 2022-04-05 | 36417532 | - | - | - | - | N/A | - | - | Spike glycoprotein,Fibritin | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7zh5 | 3.30 Å | 2023-02-15 | Cryo-EM | NAG | SARS-CoV | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-04-05 | 36417532 | - | - | - | - | N/A | - | - | Spike glycoprotein,Fibritin | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7y7j | 4.80 Å | 2023-02-22 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-06-22 | 36726762 | - | - | - | - | N/A | - | - | 1F VL; Spike glycoprotein; 1F VH | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7y7k | 4.40 Å | 2023-02-22 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-06-22 | 36726762 | - | - | - | - | N/A | - | - | Spike protein S1; 1F VL; 1F VH | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8b2t | 1.89 Å | 2023-02-22 | X-ray | OW1 | SARS-CoV-2 | NSP5 (3CLpro) | 18.24% | 22.34% | 54.30% | Nirmatrelvir (reacted form) | Nirmatrelvir (reacted form) | xia2.multiplex | 79.10% | 7.6 | Diamond (VMXm) | REFMAC; PDB-REDO | 2022-09-14 | 36757959 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 61.8 | - | - | - | |||||||
| 8c8p | 4.10 Å | 2023-02-22 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2023-01-20 | 36895554 | - | - | - | - | N/A | - | - | Spike glycoprotein,SARS-CoV-2 spike glycoprotein; Heavy-chain-only antibody 10D12 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8g62 | 2.17 Å | 2023-02-22 | X-ray | YOO, NA | SARS-CoV-2 | NSP3: PLpro | 17.96% | 21.45% | 14.30% | 3-methoxy-5-(1-methylpiperidin-4-yl)-N-[4-(pyrrolidine-1-sulfonyl)phenyl]benzamide; SODIUM ION | 3-methoxy-5-(1-methylpiperidin-4-yl)-N-[4-(pyrrolidine-1-sulfonyl)phenyl]benzamide; SODIUM ION | HKL-3000 | 100.00% | 7.1 | APS (19-ID) | REFMAC | 2023-02-14 | - | ZN; NA | - | - | - | N/A | - | - | Non-structural protein 3 | - | - | No | - | - | 70.3 | - | - | - | ||||||
| 7u0q | 3.86 Å | 2023-03-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-18 | 37167972 | - | - | - | - | N/A | - | - | mAb 002-02 light chain; mAb 002-02 Heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7u0x | 3.82 Å | 2023-03-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-19 | 37167972 | - | - | - | - | N/A | - | - | mAb 002-13 Light chain; Spike glycoprotein; mAb 002-13 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wp6 | 3.81 Å | 2023-03-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-23 | 36323313 | - | - | - | - | N/A | - | - | 85F7 heavy chain; 85F7 light chain; 36H6 light chain; Spike protein S1; 83H7 heavy chain; 83H7 light chain; 36H6 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wsc | 3.78 Å | 2023-03-01 | Cryo-EM | - | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-01-28 | - | - | - | - | - | N/A | - | - | Spike protein S1 - RBD; 3500H; 3500L | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||||
| 7x2h | 2.10 Å | 2023-03-01 | X-ray | NAG | SARS-CoV-2 | Spike | 22.83% | 26.89% | 98.90% | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.80% | 5.7 | SSRF (BL02U1) | PHENIX | 2022-02-25 | 37069158 | - | - | - | - | N/A | - | - | Spike protein S1 - receptor-binding domain; 6-2C L chain; 6-2C H chain | - | - | No | - | 20.3 | - | - | - | ||||||||
| 7xcz | 3.20 Å | 2023-03-01 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-03-26 | 36609558 | - | - | - | - | N/A | - | - | BA7054 fab; BA7125 fab; BA7125 fab; BA7054 fab; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7xda | 2.98 Å | 2023-03-01 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-03-26 | 36609558 | - | - | - | - | N/A | - | - | Spike glycoprotein; BA7208 fab; BA7208 fab; BA7125 fab; BA7125 fab | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7xdb | 2.62 Å | 2023-03-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-03-26 | 36609558 | - | - | - | - | N/A | - | - | BA7208 fab; Spike glycoprotein; BA7208 fab | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7xdk | 3.20 Å | 2023-03-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-03-27 | 36609558 | - | - | - | - | N/A | - | - | BA7054 fab; Spike glycoprotein; BA7125 fab; BA7125 fab; BA7054 fab | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7xq6 | 2.00 Å | 2023-03-01 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 18.81% | 23.26% | N/A | - | XDS | 99.20% | 16.2 | RRCAT INDUS-2 (PX-BL21) | PHENIX | 2022-05-06 | 36650888 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 52.5 | - | - | - | ||||||||
| 7xq7 | 2.35 Å | 2023-03-01 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 20.86% | 23.85% | N/A | SODIUM ION | SODIUM ION | XDS | 99.80% | 10.6 | RRCAT INDUS-2 (PX-BL21) | PHENIX | 2022-05-06 | 36650888 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 46.4 | - | - | - | |||||||
| 7zsd | 3.29 Å | 2023-03-01 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-05-06 | 37100904 | - | - | - | - | N/A | - | - | de novo designed binder; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7zss | 2.63 Å | 2023-03-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-05-08 | 37100904 | - | - | - | - | N/A | - | - | de novo designed binder; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8aqs | 2.92 Å | 2023-03-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-08-13 | 37018380 | ZN | - | - | - | N/A | - | - | Spike glycoprotein,Fibritin; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8aqt | 4.40 Å | 2023-03-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-08-13 | 37018380 | - | - | - | - | N/A | - | - | Spike glycoprotein,Fibritin; Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, membrane-bound form | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8aqu | 3.22 Å | 2023-03-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-08-13 | 37018380 | - | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, membrane-bound form; Spike glycoprotein,Fibritin | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8aqv | 2.96 Å | 2023-03-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-08-13 | 37018380 | - | - | - | - | N/A | - | - | Spike glycoprotein,Fibritin; Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, membrane-bound form | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8c5m | 1.90 Å | 2023-03-01 | X-ray | MTA, MES | SARS-CoV-2 | NSP10/NSP16 | 18.36% | 21.12% | 15.31% | 5'-DEOXY-5'-METHYLTHIOADENOSINE; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | 5'-DEOXY-5'-METHYLTHIOADENOSINE; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | XDS | 99.63% | 13.34 | PETRA III, EMBL c/o DESY (P13 (MX1)) | PHENIX | 2023-01-09 | - | ZN | - | - | - | N/A | - | - | 2'-O-methyltransferase nsp16; Non-structural protein 10 | - | - | No | - | - | 73.0 | - | - | - | ||||||
| 8d35 | 1.90 Å | 2023-03-01 | X-ray | - | SARS-CoV-2 | NSP5/RNA/RNA | 18.88% | 22.44% | N/A | tRNA (guanine(26)-N(2))-dimethyltransferase; SODIUM ION | SODIUM ION | XDS | 99.08% | 9.7 | NSLS-II (17-ID-1) | PHENIX | 2022-05-31 | 36865253 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 - UNP residues 3264-3569; tRNA (guanine(26)-N(2))-dimethyltransferase - UNP residues 526-536 | - | - | No | - | 60.8 | - | - | tRNA (guanine(26)-N(2))-dimethyltransferase | - | ||||||
| 7ul0 | 2.49 Å | 2023-03-08 | X-ray | NAG, NA | SARS-CoV-2 | Spike | 22.46% | 25.59% | 14.40% | 2-acetamido-2-deoxy-beta-D-glucopyranose; SODIUM ION | 2-acetamido-2-deoxy-beta-D-glucopyranose; SODIUM ION | HKL-3000 | 99.90% | 5.3 | APS (23-ID-B) | PHENIX | 2022-04-03 | 36514310 | - | - | - | - | N/A | - | - | Heavy chain of EH8; Light chain of EH8; Spike protein S1 | - | - | No | - | 30.2 | - | - | - | |||||||
| 7ul1 | 2.65 Å | 2023-03-08 | X-ray | NAG | Spike | 17.40% | 22.06% | 19.00% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-3000 | 99.40% | 3.7 | SSRL (BL9-2) | PHENIX | 2022-04-03 | 36514310 | - | - | - | - | N/A | - | - | Spike protein S1; Light chain of EH3; Heavy chain of EH3 | - | - | No | - | 64.6 | - | - | - | ||||||||
| 7wp8 | 3.88 Å | 2023-03-08 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-01-23 | 36323313 | - | - | - | - | N/A | - | - | 83H7 heavy chain; 2B4 heavy chain; Spike glycoprotein; 2B4 light chain; 85F7 heavy chain; 85F7 light chain; 83H7 light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7x63 | 2.24 Å | 2023-03-08 | X-ray | - | SARS-CoV-2 | Spike | 19.27% | 22.43% | 12.00% | - | HKL-2000 | 99.50% | 4.2 | SSRF (BL19U1) | PHENIX | 2022-03-06 | - | - | - | - | - | N/A | - | - | BD-236 Fab heavy chain; Spike protein S1; BD-236 Fab light chain | - | - | No | - | - | 60.9 | - | - | - | |||||||
| 7x66 | 2.40 Å | 2023-03-08 | X-ray | - | SARS-CoV-2 | Spike | 18.99% | 23.82% | 18.30% | - | HKL-2000 | 99.10% | 2.9 | SSRF (BL19U1) | PHENIX | 2022-03-06 | - | - | - | - | - | N/A | - | - | BD-236 Fab light chain; BD-236 Fab heavy chain; Spike protein S1 | - | - | No | - | - | 46.6 | - | - | - | |||||||
| 7zrv | 2.80 Å | 2023-03-08 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-05-05 | 37100904 | - | - | - | - | N/A | - | - | de novo designed binder; Spike glycoprotein,Envelope glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8bev | 5.92 Å | 2023-03-08 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-10-21 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; immunoglobulin mu heavy chain | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 8bgg | 6.04 Å | 2023-03-08 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-10-27 | - | - | - | - | - | N/A | - | - | Nanobody W25; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 8c19 | 1.95 Å | 2023-03-08 | X-ray | T6B | SARS-CoV-2 | NSP3: Macro | 17.94% | 22.62% | N/A | [5-(1-methylpyrrolo[2,3-b]pyridin-4-yl)furan-2-yl]-morpholin-4-yl-methanone | [5-(1-methylpyrrolo[2,3-b]pyridin-4-yl)furan-2-yl]-morpholin-4-yl-methanone | DIALS | 100.00% | 5.5 | Diamond (I03) | REFMAC | 2022-12-20 | 36839595 | - | - | - | - | N/A | - | - | Non-structural protein 3 | - | - | No | - | 58.9 | - | - | - | |||||||
| 8c1a | 1.90 Å | 2023-03-08 | X-ray | T6O | SARS-CoV-2 | NSP3: Macro | 17.05% | 21.41% | N/A | aztreonam | aztreonam | DIALS | 95.20% | 62.9 | Diamond (I03) | REFMAC | 2022-12-20 | 36839595 | - | - | - | - | N/A | - | - | Replicase polyprotein 1ab | - | - | No | - | 70.5 | - | - | - | |||||||
| 8f2e | 2.43 Å | 2023-03-08 | X-ray | - | SARS-CoV-2 | NSP3(C-term) | 19.00% | 23.20% | N/A | - | autoPROC | 97.50% | 31.6 | NSLS-II, SSRL (17-ID-2, BL12-2) | BUSTER | 2022-11-07 | 36801935 | - | - | - | - | N/A | - | - | Papain-like protease nsp3 - CoV-Y domain | - | - | No | - | 52.9 | - | - | - | ||||||||
| 7u8e | 2.29 Å | 2023-03-15 | X-ray | NAG | SARS-CoV-2 | Spike | 20.40% | 24.75% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 96.30% | 9.78 | APS (19-ID) | PHENIX | 2022-03-08 | - | - | - | - | - | N/A | - | - | Antibody Ab246 Fab light chain; Spike protein S1 - receptor binding domain (RBD); Antibody Ab246 Fab heavy chain | - | - | No | - | - | 37.7 | - | - | - | ||||||
| 7xdl | 3.08 Å | 2023-03-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-03-27 | 36609558 | - | - | - | - | N/A | - | - | BA7125 fab; BA7208 fab; BA7208 fab; Spike glycoprotein; BA7125 fab | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8aeb | 1.83 Å | 2023-03-15 | X-ray | NA, 35J | SARS-CoV-2 | NSP5 (3CLpro) | 18.35% | 23.41% | 9.30% | SODIUM ION; N-(pyridin-3-ylmethyl)thioformamide | SODIUM ION; N-(pyridin-3-ylmethyl)thioformamide | XDS | 98.70% | 9.9 | SOLEIL (PROXIMA 2) | REFMAC; REFMAC | 2022-07-12 | 36796300 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 50.8 | - | - | - | |||||||
| 8aqw | 3.30 Å | 2023-03-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-08-13 | 37018380 | - | - | - | - | N/A | - | - | Spike glycoprotein,Fibritin; Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, A allele | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wqa | 1.80 Å | 2023-03-22 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 17.12% | 20.76% | 4.10% | Z-VAD(OMe)-FMK | - | HKL-2000 | 99.40% | 29.568 | NSRRC (BL15A1) | REFMAC | 2022-01-25 | - | - | - | - | - | N/A | - | - | 3C-like proteinase; Z-VAD(OMe)-FMK | - | - | No | - | - | 76.1 | - | - | - | ||||||
| 7x27 | 2.49 Å | 2023-03-22 | Cryo-EM | - | MERS-CoV | Spike | N/A | N/A | N/A | - | - | - | () | 2022-02-25 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||||
| 7x93 | 3.30 Å | 2023-03-22 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-03-15 | 37288342 | - | - | - | - | N/A | - | - | Ab765 light chain; Spike glycoprotein; Ab765 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7x94 | 4.00 Å | 2023-03-22 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | phenix.real_space_refine; PHENIX | 2022-03-15 | 37288342 | - | - | - | - | N/A | - | - | Ab712 light chain; Spike glycoprotein; Ab712 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7x95 | 3.90 Å | 2023-03-22 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX; phenix.real_space_refine | 2022-03-15 | 37288342 | - | - | - | - | N/A | - | - | Ab709 light chain; Spike glycoprotein; Ab709 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7x96 | 3.40 Å | 2023-03-22 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX; phenix.real_space_refine | 2022-03-15 | 37288342 | - | - | - | - | N/A | - | - | Ab847 light chain; Spike glycoprotein; Ab847 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7xax | 2.25 Å | 2023-03-22 | X-ray | 3WL | SARS-CoV-2 | NSP5 (3CLpro) | 22.89% | 27.97% | 7.10% | 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one | 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one | 97.70% | 11.0 | SSRF (BL02U1) | PHENIX | 2022-03-19 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 14.3 | - | - | - | |||||||
| 7xb3 | 2.08 Å | 2023-03-22 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 21.36% | 24.25% | 3.10% | - | XDS | 100.00% | 18.5 | SSRF (BL02U1) | PHENIX | 2022-03-20 | - | - | - | - | - | N/A | - | - | Replicase polyprotein 1a | - | - | No | - | - | 42.6 | - | - | - | |||||||
| 7xb4 | 2.07 Å | 2023-03-22 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | 21.09% | 24.03% | 3.70% | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | XDS | 94.40% | 15.7 | SSRF (BL02U1) | PHENIX | 2022-03-20 | - | - | - | - | - | N/A | - | - | Replicase polyprotein 1a | - | - | No | - | - | 44.5 | - | - | - | ||||||
| 7z2k | 1.65 Å | 2023-03-22 | X-ray | NA, MLI | SARS-CoV-2 | NSP5 (3CLpro) | 18.84% | 20.95% | 9.08% | SODIUM ION; MALONATE ION | SODIUM ION; MALONATE ION | XDS | 99.34% | 13.35 | PETRA III, DESY (P11) | PHENIX; PHENIX | 2022-02-28 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 74.5 | - | - | - | ||||||
| 7z3u | 1.72 Å | 2023-03-22 | X-ray | RN2, NA | SARS-CoV-2 | NSP5 (3CLpro) | 18.65% | 21.56% | 10.10% | Calpeptin; SODIUM ION | Calpeptin; SODIUM ION | XDS | 99.70% | 12.54 | PETRA III, DESY (P11) | PHENIX | 2022-03-02 | 37853179 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 69.3 | - | - | - | |||||||
| 7z4s | 1.70 Å | 2023-03-22 | X-ray | AEA, II7, PGE | SARS-CoV-2 | NSP5 (3CLpro) | 19.82% | 23.38% | 12.90% | DTY-PHE-HIS-LEU-ASN-LEU-GLY-TYR-ARG-PRO-GLY; 3-azanylcyclobutane-1-carbaldehyde; (2-AMINO-2-CARBAMOYL-ETHYLSULFANYL)-ACETIC ACID; TRIETHYLENE GLYCOL | (2-AMINO-2-CARBAMOYL-ETHYLSULFANYL)-ACETIC ACID; 3-azanylcyclobutane-1-carbaldehyde; TRIETHYLENE GLYCOL | xia2 | 99.90% | 6.6 | Diamond (I03) | REFMAC | 2022-03-04 | 37217786 | - | - | - | - | N/A | - | - | ORF1a polyprotein; ORF1a polyprotein; DTY-PHE-HIS-LEU-ASN-LEU-GLY-TYR-ARG-PRO-GLY | - | - | No | - | 51.4 | - | - | - | |||||||
| 8bbn | 3.58 Å | 2023-03-22 | X-ray | - | SARS-CoV-2 | Spike | 26.54% | 31.39% | 46.70% | - | xia2 | 100.00% | 4.1 | Diamond (I03) | PHENIX | 2022-10-14 | 36995936 | - | - | - | - | N/A | - | - | BA.2-10 light chain; EY6A Heavy chain; EY6A light chain; Spike protein S1; BA.2-10 heavy chain | - | - | No | - | 5.5 | - | - | - | ||||||||
| 8bbo | 2.75 Å | 2023-03-22 | X-ray | NAG | SARS-CoV-2 | Spike | 22.42% | 25.20% | 46.00% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | xia2 | 100.00% | 7.5 | Diamond (I03) | PHENIX | 2022-10-14 | 36995936 | - | - | - | - | N/A | - | - | Spike glycoprotein; Immunoglobulin kappa light chain; IGH@ protein | - | - | No | - | 33.3 | - | - | - | |||||||
| 8bcz | 2.90 Å | 2023-03-22 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-10-17 | 36995936 | - | - | - | - | N/A | - | - | Spike protein S1; BA.2-36 light chain; BA.2-36 heavy chain; EY6A light chain; EY6A heavy chain; COVOX-45 light chain; COVOX-45 heavy chain; BA.2-23 light chain; BA.2-23 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8c3v | 2.74 Å | 2023-03-22 | X-ray | PG0, PG4 | SARS-CoV-2 | Spike | 21.56% | 25.77% | 25.70% | 2-(2-METHOXYETHOXY)ETHANOL | 2-(2-METHOXYETHOXY)ETHANOL; TETRAETHYLENE GLYCOL | xia2 | 100.00% | 7.4 | Diamond (I04) | PHENIX | 2022-12-28 | 36995936 | - | - | - | - | N/A | - | - | Nanobody C1; Spike protein S1; BA.2-13 light chain; BA.2-13 heavy chain | - | - | No | - | 28.6 | - | - | - | |||||||
| 8d0z | 3.70 Å | 2023-03-22 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-05-26 | 36862518 | - | - | - | - | N/A | - | - | Spike glycoprotein; S728-1157 Fab heavy chain variable region; S728-1157 Fab light chain variable region | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8h3d | 3.27 Å | 2023-03-22 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-10-08 | 36844485 | - | - | - | - | N/A | - | - | Spike glycoprotein,Fibritin - SARS-CoV-2 spike protein,SARS-CoV-2 spike protein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8h3e | 3.06 Å | 2023-03-22 | Cryo-EM | NAG, Q83 | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; 7-(6-nitro-2,3-dihydroindol-1-yl)-7-oxidanylidene-heptanoic acid | 2-acetamido-2-deoxy-beta-D-glucopyranose; 7-(6-nitro-2,3-dihydroindol-1-yl)-7-oxidanylidene-heptanoic acid | - | - | () | 2022-10-08 | 36844485 | - | - | - | - | N/A | - | - | Spike glycoprotein,Fibritin - SARS-CoV-2 spike protein,SARS-CoV-2 spike protein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8dsu | 1.86 Å | 2023-03-29 | X-ray | V2M | SARS-CoV-2 | NSP5 (3CLpro) | 20.49% | 25.43% | 4.74% | N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | CrysalisPro | 99.90% | 19.97 | ROTATING ANODE () | PHENIX; PHENIX | 2022-07-22 | 36992489 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 31.4 | - | - | - | |||||||
| 8e7c | 2.45 Å | 2023-03-29 | X-ray | V2M | HKU-15 | NSP5 (3CLpro) | 19.27% | 23.49% | 16.08% | N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | CrysalisPro | 93.51% | 5.37 | ROTATING ANODE () | PHENIX | 2022-08-23 | 36992489 | - | - | - | - | N/A | - | - | Main Protease | - | - | No | - | 50.2 | - | - | - | |||||||
| 8e7n | 1.65 Å | 2023-03-29 | X-ray | B1S, K36 | Whale-CoV SW1 | NSP5 (3CLpro) | 15.93% | 19.50% | 5.57% | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid; (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | CrysalisPro | 94.95% | 11.31 | ROTATING ANODE () | PHENIX | 2022-08-24 | 36992489 | - | - | - | - | N/A | - | - | main protease | - | - | No | - | 84.9 | - | - | - | |||||||
| 8fa1 | 2.51 Å | 2023-03-29 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2022-11-25 | 36940324 | - | - | - | - | N/A | - | - | Spike protein S2' HR2; Ferritin, Dps family protein and Spike protein S2' chimera | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 8fa2 | 2.82 Å | 2023-03-29 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2022-11-25 | 36940324 | - | - | - | - | N/A | - | - | Scaffolded Spike protein S2' HR1; Spike protein S2' 42G | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 8fwx | 2.12 Å | 2023-03-29 | X-ray | - | Whale-CoV SW1 | NSP5 (3CLpro) | 19.86% | 23.83% | 13.50% | - | autoPROC | 99.90% | 11.3 | APS (19-ID) | PHENIX | 2023-01-23 | 36992489 | - | - | - | - | N/A | - | - | Main Protease | - | - | No | - | 46.5 | - | - | - | ||||||||
| 8g6r | 3.30 Å | 2023-03-29 | Cryo-EM | - | PEDV | NSP7/NSP8/NSP12/Spike | N/A | N/A | N/A | RNA (5'-R(P*GP*GP*UP*UP*GP*UP*GP*AP*UP*UP*UP*UP*AP*AP*UP*AP*GP*CP*UP*U)-3'); RNA (5'-R(P*AP*AP*GP*AP*AP*GP*CP*UP*AP*UP*UP*AP*AP*AP*AP*UP*CP*AP*CP*A)-3') | - | - | - | () | PHENIX | 2023-02-15 | 36993498 | ZN | - | - | - | N/A | - | - | nsp8; nsp12; RNA (5'-R(P*AP*AP*GP*AP*AP*GP*CP*UP*AP*UP*UP*AP*AP*AP*AP*UP*CP*AP*CP*A)-3'); RNA (5'-R(P*GP*GP*UP*UP*GP*UP*GP*AP*UP*UP*UP*UP*AP*AP*UP*AP*GP*CP*UP*U)-3'); nsp7 | N/A | - | No | - | N/A | N/A | N/A | RNA (5'-R(P*GP*GP*UP*UP*GP*UP*GP*AP*UP*UP*UP*UP*AP*AP*UP*AP*GP*CP*UP*U)-3'); RNA (5'-R(P*AP*AP*GP*AP*AP*GP*CP*UP*AP*UP*UP*AP*AP*AP*AP*UP*CP*AP*CP*A)-3') | - | |||||||
| 8hht | 1.95 Å | 2023-03-29 | X-ray | LV0 | SARS-CoV-2 | NSP5 (3CLpro) | 19.64% | 24.50% | 4.60% | ~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(1,3-thiazol-2-ylmethylamino)butan-2-yl]benzamide | ~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(1,3-thiazol-2-ylmethylamino)butan-2-yl]benzamide | XDS | 87.30% | 23.3 | SSRF (BL18U1) | PHENIX | 2022-11-17 | 36928316 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 40.0 | - | - | - | |||||||
| 8hhu | 2.26 Å | 2023-03-29 | X-ray | LVX | SARS-CoV-2 | NSP5 (3CLpro) | 18.54% | 22.68% | 6.70% | (1~{R})-3,3-bis(fluoranyl)-~{N}-[(2~{R})-3-methoxy-1-oxidanylidene-1-[[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(1,3-thiazol-2-ylmethylamino)butan-2-yl]amino]propan-2-yl]cyclohexane-1-carboxamide | (1~{R})-3,3-bis(fluoranyl)-~{N}-[(2~{R})-3-methoxy-1-oxidanylidene-1-[[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(1,3-thiazol-2-ylmethylamino)butan-2-yl]amino]propan-2-yl]cyclohexane-1-carboxamide | XDS | 98.80% | 14.0 | MAX IV (BioMAX) | BUSTER | 2022-11-17 | 36928316 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 58.4 | - | - | - | |||||||
| 8dzb | 1.85 Å | 2023-04-05 | X-ray | U6Y | SARS-CoV-2 | NSP5 (3CLpro) | 18.33% | 20.96% | 8.70% | benzyl {(3S)-1-[(2S)-1-({(2S,3R)-4-(cyclopropylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-5-oxopyrrolidin-3-yl}carbamate | benzyl {(3S)-1-[(2S)-1-({(2S,3R)-4-(cyclopropylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-5-oxopyrrolidin-3-yl}carbamate | iMOSFLM | 94.70% | 9.8 | APS (22-ID) | REFMAC | 2022-08-06 | 36920943 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 74.4 | - | - | - | |||||||
| 8dzc | 2.20 Å | 2023-04-05 | X-ray | U76 | SARS-CoV-2 | NSP5 (3CLpro) | 21.48% | 24.47% | 6.00% | (3,5-difluorophenyl)methyl {(3S)-1-[(2S)-1-({(2S,3R)-4-(cyclopropylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-6-oxopiperidin-3-yl}carbamate | (3,5-difluorophenyl)methyl {(3S)-1-[(2S)-1-({(2S,3R)-4-(cyclopropylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-6-oxopiperidin-3-yl}carbamate | iMOSFLM | 97.80% | 15.7 | APS (22-ID) | REFMAC | 2022-08-06 | 36920943 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 40.5 | - | - | - | |||||||
| 8eua | 3.10 Å | 2023-04-05 | X-ray | WUK | SARS-CoV-2 | NSP3: PLpro | 19.13% | 25.34% | 43.60% | methyl 4-{2-[3-(2-{[(1R)-1-(naphthalen-1-yl)ethyl]carbamoyl}phenyl)propanoyl]hydrazinyl}-4-oxobutanoate | methyl 4-{2-[3-(2-{[(1R)-1-(naphthalen-1-yl)ethyl]carbamoyl}phenyl)propanoyl]hydrazinyl}-4-oxobutanoate | XDS | 99.80% | 10.91 | SSRL (BL12-2) | REFMAC | 2022-10-18 | 36977673 | ZN | - | - | - | N/A | - | - | Papain-like protease nsp3 | - | - | No | - | 32.2 | - | - | - | |||||||
| 8fu7 | 3.21 Å | 2023-04-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2023-01-16 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 8fu8 | 3.08 Å | 2023-04-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2023-01-16 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 8fu9 | 3.52 Å | 2023-04-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2023-01-16 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 8gjm | 2.80 Å | 2023-04-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2023-03-16 | 36916782 | - | - | - | - | N/A | - | - | Heavy chain of 17B10 fab; Light chain of 17B10 fab; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8gjn | 3.60 Å | 2023-04-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2023-03-16 | 36916782 | - | - | - | - | N/A | - | - | Spike protein S2' - RBD; Heavy chain of 17B10 Fab; Light chain of 17B10 Fab | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8ign | 2.02 Å | 2023-04-05 | X-ray | 7ON | SARS-CoV-2 | NSP5 (3CLpro) | 18.62% | 23.12% | 6.70% | (3~{S},3~{a}~{S},6~{a}~{R})-2-[(2~{S})-2-cyclohexyl-2-[2,2,2-tris(fluoranyl)ethanoylamino]ethanoyl]-~{N}-[(2~{S})-4-(cyclopentylamino)-3,4-bis(oxidanylidene)-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide | (3~{S},3~{a}~{S},6~{a}~{R})-2-[(2~{S})-2-cyclohexyl-2-[2,2,2-tris(fluoranyl)ethanoylamino]ethanoyl]-~{N}-[(2~{S})-4-(cyclopentylamino)-3,4-bis(oxidanylidene)-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide | XDS | 97.50% | 9.9 | SSRF (BL19U1) | REFMAC | 2023-02-21 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 53.8 | - | - | - | ||||||
| 8igo | 2.00 Å | 2023-04-05 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 20.59% | 24.90% | 3.10% | - | XDS | 96.20% | 31.6 | SSRF (BL19U1) | PHENIX | 2023-02-21 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 36.0 | - | - | - | |||||||
| 7sh7 | 1.85 Å | 2023-04-12 | X-ray | 9GI | SARS-CoV-2 | NSP5 (3CLpro) | 25.78% | 29.72% | 8.90% | benzyl [(2S,3R)-3-tert-butoxy-1-{[(2S)-3-cyclohexyl-1-oxo-1-(2-{[(3S)-2-oxopyrrolidin-3-yl]methyl}-2-propanoylhydrazinyl)propan-2-yl]amino}-1-oxobutan-2-yl]carbamate (non-preferred name) | benzyl [(2S,3R)-3-tert-butoxy-1-{[(2S)-3-cyclohexyl-1-oxo-1-(2-{[(3S)-2-oxopyrrolidin-3-yl]methyl}-2-propanoylhydrazinyl)propan-2-yl]amino}-1-oxobutan-2-yl]carbamate (non-preferred name) | PROTEUM | 97.00% | 9.3 | ROTATING ANODE () | PHENIX | 2021-10-08 | 37090597 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 7.9 | - | - | - | |||||||
| 7sh8 | 1.80 Å | 2023-04-12 | X-ray | GJ3 | SARS-CoV-2 | NSP5 (3CLpro) | 23.45% | 27.24% | N/A | benzyl [(2S,3R)-1-{[(2S)-1-(2-acetyl-2-{[(3S)-2-oxopyrrolidin-3-yl]methyl}hydrazinyl)-3-cyclohexyl-1-oxopropan-2-yl]amino}-3-tert-butoxy-1-oxobutan-2-yl]carbamate (non-preferred name) | benzyl [(2S,3R)-1-{[(2S)-1-(2-acetyl-2-{[(3S)-2-oxopyrrolidin-3-yl]methyl}hydrazinyl)-3-cyclohexyl-1-oxopropan-2-yl]amino}-3-tert-butoxy-1-oxobutan-2-yl]carbamate (non-preferred name) | PROTEUM PLUS | 99.60% | 12.9 | ROTATING ANODE () | PHENIX | 2021-10-08 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 18.2 | - | - | - | ||||||
| 7sh9 | 1.85 Å | 2023-04-12 | X-ray | 9HA | SARS-CoV-2 | NSP5 (3CLpro) | 22.58% | 26.30% | N/A | benzyl [(2S,3R)-1-({(2S)-1-[2-acetyl-2-(3-amino-3-oxopropyl)hydrazinyl]-3-cyclohexyl-1-oxopropan-2-yl}amino)-3-tert-butoxy-1-oxobutan-2-yl]carbamate (non-preferred name) | benzyl [(2S,3R)-1-({(2S)-1-[2-acetyl-2-(3-amino-3-oxopropyl)hydrazinyl]-3-cyclohexyl-1-oxopropan-2-yl}amino)-3-tert-butoxy-1-oxobutan-2-yl]carbamate (non-preferred name) | PROTEUM PLUS | 99.60% | 8.9 | ROTATING ANODE () | PHENIX | 2021-10-08 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 24.3 | - | - | - | ||||||
| 8fez | 3.72 Å | 2023-04-12 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2022-12-07 | 36993551 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 8gzz | 3.52 Å | 2023-04-12 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-09-27 | - | - | - | - | - | N/A | - | - | Spike protein S1 - SARS-CoV-2 Omicron BA.1 Spike glycoprotein receptor binding domain; rabbit monoclonal antibody 1H1 Fab heavy chain; rabbit monoclonal antibody 1H1 Fab light chain | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 8h00 | 3.41 Å | 2023-04-12 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-09-27 | - | - | - | - | - | N/A | - | - | rabbit monoclonal antibody 1H1 Fab heavy chain; rabbit monoclonal antibody 1H1 Fab light chain; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 8h01 | 3.70 Å | 2023-04-12 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-09-27 | - | - | - | - | - | N/A | - | - | rabbit monoclonal antibody 1H1 Fab heavy chain; rabbit monoclonal antibody 1H1 Fab light chain; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 8itu | 3.68 Å | 2023-04-12 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2023-03-23 | - | - | - | - | - | N/A | - | - | 1H1 heavy chain; 1H1 light chain; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7uow | 4.40 Å | 2023-04-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-04-14 | 37167972 | - | - | - | - | N/A | - | - | Spike glycoprotein; Monoclonal antibody 034_32 heavy chain; Monoclonal antibody 034_32 light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wn2 | 2.35 Å | 2023-04-19 | X-ray | NAG | SARS-CoV-2 | Spike | 22.82% | 27.85% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.60% | 4.1 | SPring-8 (BL44XU) | PHENIX | 2022-01-17 | 37041231 | - | - | - | - | N/A | - | - | Fab Heavy chain; Fab Light chain; Spike protein S1 - Receptor-binding domain | - | - | No | - | 14.8 | - | - | - | ||||||||
| 7wnb | 2.18 Å | 2023-04-19 | X-ray | - | SARS-CoV-2 | Spike | 24.49% | 28.07% | N/A | - | XDS | 99.80% | 5.1 | SPring-8 (BL44XU) | PHENIX | 2022-01-18 | 37041231 | - | - | - | - | N/A | - | - | Spike protein S1 - Receptor-binding domain; Fab Light chain; Fab Heavy chain | - | - | No | - | 13.8 | - | - | - | ||||||||
| 7xj6 | 3.29 Å | 2023-04-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-04-15 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; 58G6 heavy chain; 58G6 light chain; 55A8 heavy chain; 55A8 light chain | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7xj8 | 3.30 Å | 2023-04-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-04-15 | - | - | - | - | - | N/A | - | - | 55A8 heavy chain; 58G6 light chain; 58G6 heavy chain; 55A8 light chain; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7xj9 | 3.27 Å | 2023-04-19 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-04-15 | - | - | - | - | - | N/A | - | - | 58G6 light chain; 55A8 light chain; 58G6 heavy chain; 55A8 heavy chain; Spike glycoprotein - UNP residues 334-527 | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7yow | 3.30 Å | 2023-04-19 | X-ray | - | SARS-CoV-2 | Spike | 20.85% | 25.42% | 9.40% | - | XDS | 97.80% | 5.5 | SPring-8 (BL44XU) | PHENIX | 2022-08-02 | 37041231 | - | - | - | - | N/A | - | - | Fab Heavy chain; Fab Light chain; Spike protein S1 | - | - | No | - | 31.4 | - | - | - | ||||||||
| 7zjl | 2.60 Å | 2023-04-19 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2022-04-11 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; REGN10987 Fab homologue (Light chain); REGN10987 Fab homologue (Heavy chain) | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||||
| 8c1v | 2.90 Å | 2023-04-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-12-21 | 36964125 | - | - | - | - | N/A | - | - | Spike glycoprotein; Sb92 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8hef | 1.51 Å | 2023-04-19 | X-ray | 7YY | SARS-CoV-2 | NSP5 (3CLpro) | 13.26% | 17.01% | 7.10% | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | xia2 | 99.80% | 14.7 | SSRF (BL19U1) | REFMAC | 2022-11-08 | 36997073 | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | 95.1 | - | - | - | ||||||||
| 8i5h | 2.38 Å | 2023-04-19 | X-ray | NAG | SARS-CoV-2 | Spike | 18.83% | 23.03% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 100.00% | 10.3 | SPring-8 (BL44XU) | PHENIX | 2023-01-25 | 37041231 | - | - | - | - | N/A | - | - | Spike protein S1 - Delta RBD; Fab Light chain; Fab Heavy chain | - | - | No | - | 54.7 | - | - | - | ||||||||
| 8i5i | 3.06 Å | 2023-04-19 | X-ray | - | SARS-CoV-2 | Spike | 22.83% | 37.25% | N/A | - | XDS | 88.60% | 8.9 | SPring-8 (BL44XU) | PHENIX | 2023-01-25 | 37041231 | - | - | - | - | N/A | - | - | Fab Light chain; Fab Heavy chain; Spike protein S1 | - | - | No | - | 3.6 | - | - | - | ||||||||
| 5sml | 1.53 Å | 2023-04-26 | X-ray | O3R | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.24% | 22.73% | 8.80% | 6-{[(3,4-dichlorophenyl)methyl](methyl)amino}pyridine-3-sulfonamide | 6-{[(3,4-dichlorophenyl)methyl](methyl)amino}pyridine-3-sulfonamide | XDS | 99.60% | 11.2 | Diamond (I04-1) | REFMAC | 2022-04-20 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 57.8 | - | - | - | |||||
| 5smm | 1.58 Å | 2023-04-26 | X-ray | O46 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.06% | 23.11% | 9.70% | N-[4-(3-fluorophenyl)oxan-4-yl]-2-(3-hydroxyphenyl)acetamide | N-[4-(3-fluorophenyl)oxan-4-yl]-2-(3-hydroxyphenyl)acetamide | XDS | 99.70% | 12.2 | Diamond (I04-1) | REFMAC | 2022-04-20 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 53.9 | - | - | - | |||||
| 5smn | 1.36 Å | 2023-04-26 | X-ray | O4F | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.73% | 21.16% | 6.80% | N-(1-cyanocyclopropyl)-1-(3-methylpyridin-4-yl)piperidine-4-carboxamide | N-(1-cyanocyclopropyl)-1-(3-methylpyridin-4-yl)piperidine-4-carboxamide | XDS | 98.80% | 14.6 | Diamond (I04-1) | REFMAC | 2022-04-20 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 72.7 | - | - | - | |||||
| 7xik | 2.89 Å | 2023-04-26 | X-ray | - | SARS-CoV-2 | Spike | 21.06% | 26.30% | 26.20% | - | DENZO | 99.80% | 10.2 | SSRF (BL19U1) | PHENIX | 2022-04-13 | - | - | - | - | - | N/A | - | - | B38 Fab heavy chain; Spike protein S1; B38 Fab light chain | - | - | No | - | - | 24.3 | - | - | - | |||||||
| 7xil | 2.91 Å | 2023-04-26 | X-ray | NAG | SARS-CoV-2 | Spike | 20.63% | 25.62% | 19.30% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.40% | 2.4 | SSRF (BL19U1) | PHENIX | 2022-04-13 | - | - | - | - | - | N/A | - | - | Spike protein S1; B38 Fab heavy chain; B38 Fab light chain | - | - | No | - | - | 29.7 | - | - | - | ||||||
| 7xs8 | 2.80 Å | 2023-04-26 | X-ray | NAG | SARS-CoV-2 | Spike | 21.55% | 26.74% | 23.10% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 98.60% | 3.8 | SSRF (BL18U1) | PHENIX | 2022-05-13 | 36914890 | - | - | - | - | N/A | - | - | Spike protein S1; P5S-1H1 Light chain; P5S-1H1 Heavy chain | - | - | No | - | 21.3 | - | - | - | |||||||
| 8djj | 2.51 Å | 2023-04-26 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 21.79% | 27.98% | N/A | - | XDS | 97.14% | 14.96 | CLSI (08B1-1) | PHENIX | 2022-06-30 | 37122453 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 14.2 | - | - | - | ||||||||
| 8dkz | 3.00 Å | 2023-04-26 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 23.34% | 31.76% | N/A | - | XDS | 94.59% | 13.15 | CLSI (08B1-1) | PHENIX | 2022-07-06 | 37122453 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 5.2 | - | - | - | ||||||||
| 8eir | 2.49 Å | 2023-04-26 | Cryo-EM | - | SARS-CoV-2 | NSP5 (3CLpro) | N/A | N/A | N/A | - | - | - | () | phenix.real_space_refine; PHENIX | 2022-09-15 | 37044215 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5; nsp7-nsp10 of Replicase polyprotein 1a | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8ej9 | 2.50 Å | 2023-04-26 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 25.56% | 32.87% | N/A | - | XDS | 97.00% | 8.73 | SSRL (BL12-1) | PHENIX | 2022-09-16 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 4.5 | - | - | - | |||||||
| 8eke | 3.36 Å | 2023-04-26 | Cryo-EM | - | SARS-CoV-2 | NSP5 (3CLpro) | N/A | N/A | N/A | - | - | - | () | PHENIX; phenix.real_space_refine | 2022-09-20 | 37044215 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8gb0 | 4.10 Å | 2023-04-26 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2023-02-24 | 37034621 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8heb | 3.53 Å | 2023-04-26 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-11-08 | 37062240 | - | - | - | - | N/A | - | - | Spike glycoprotein; rabbit antibody 9H1 light chain; rabbit antibody 9H1 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8hec | 3.50 Å | 2023-04-26 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-11-08 | 37062240 | - | - | - | - | N/A | - | - | rabbit antibody 9H1 heavy chain; Spike glycoprotein; rabbit antibody 9H1 light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8hed | 3.59 Å | 2023-04-26 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-11-08 | 37062240 | - | - | - | - | N/A | - | - | rabbit antibody 9H1 heavy chain; Spike protein S1 - RBD; rabbit antibody 9H1 light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8a94 | 2.40 Å | 2023-05-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-06-27 | 36574772 | - | - | - | - | N/A | - | - | SARS-CoV2 Spike in 2-up conformation in complex with Fab47; Fab47 (Heavy chain Variable domain); Fab47 (Light chain Variable domain) | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8a96 | 2.70 Å | 2023-05-03 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-06-27 | 36574772 | - | - | - | - | N/A | - | - | Spike protein S1; Fab47 Heavy chain (variable domain); Fab47 Light chain (variable domain) | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8di3 | 1.50 Å | 2023-05-03 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 18.41% | 21.80% | N/A | - | XDS | 98.01% | 11.03 | SSRL (BL12-1) | PHENIX | 2022-06-28 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 66.9 | - | - | - | |||||||
| 8dkh | 1.95 Å | 2023-05-03 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 24.67% | 28.80% | N/A | - | XDS | 97.20% | 4.04 | SSRL (BL12-1) | PHENIX | 2022-07-05 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 10.6 | - | - | - | |||||||
| 8dkj | 2.11 Å | 2023-05-03 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 19.45% | 24.76% | N/A | - | XDS | 98.83% | 1.16 | SSRL (BL12-1) | PHENIX | 2022-07-05 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 37.6 | - | - | - | |||||||
| 8dkk | 2.00 Å | 2023-05-03 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 20.35% | 24.97% | N/A | - | XDS | 98.82% | 1.55 | SSRL (BL12-1) | PHENIX | 2022-07-05 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 35.6 | - | - | - | |||||||
| 8dkl | 1.90 Å | 2023-05-03 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 19.98% | 24.13% | N/A | - | XDS | 98.00% | 1.38 | SSRL (BL12-1) | PHENIX | 2022-07-05 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 43.6 | - | - | - | |||||||
| 8dmn | 2.30 Å | 2023-05-03 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 21.15% | 27.24% | N/A | - | XDS | 97.44% | 17.4 | SSRL (BL12-2) | PHENIX | 2022-07-08 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 18.2 | - | - | - | |||||||
| 8ej7 | 2.30 Å | 2023-05-03 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 20.12% | 27.02% | N/A | - | XDS | 97.08% | 17.05 | SSRL (BL12-1) | PHENIX | 2022-09-16 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 19.4 | - | - | - | |||||||
| 8f0g | 3.35 Å | 2023-05-03 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-11-02 | 37083327 | - | - | - | - | N/A | - | - | Antibody 1C3 Fab Heavy Chain; Antibody 1C3 Fab Light Chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8f0h | 3.18 Å | 2023-05-03 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | - | () | PHENIX | 2022-11-02 | 37083327 | - | - | - | - | N/A | - | - | Antibody Fab 1H2 heavy chain; Antibody Fab 2A10 light chain; Antibody Fab 2A10 heavy chain; Antibody Fab 1H2 light chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8gs9 | 2.66 Å | 2023-05-03 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-09-05 | - | - | - | - | - | N/A | - | - | Heavy chain of VacBB-551; Light chain of VacBB-551; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 8okk | 1.63 Å | 2023-05-03 | X-ray | 83F, FMT, NA, MPD | SARS-CoV-2 | NSP5 (3CLpro) | 15.22% | 17.77% | 11.20% | (4S)-2-METHYL-2,4-PENTANEDIOL; FORMIC ACID; SODIUM ION; tert-butyl-N-[(2S)-3-methyl-1-[(2S,4S)-4-methyl-2-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]carbamoyl]pyrrolidin-1-yl]-1-oxidanylidene-butan-2-yl]carbamate | tert-butyl-N-[(2S)-3-methyl-1-[(2S,4S)-4-methyl-2-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]carbamoyl]pyrrolidin-1-yl]-1-oxidanylidene-butan-2-yl]carbamate; FORMIC ACID; SODIUM ION; (4S)-2-METHYL-2,4-PENTANEDIOL | XDS | 98.00% | 13.1 | ELETTRA (11.2C) | PHENIX; PHENIX | 2023-03-28 | 37043904 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 93.0 | - | - | - | |||||||
| 8okl | 1.50 Å | 2023-05-03 | X-ray | NA, 83N, FMT, MPD | SARS-CoV-2 | NSP5 (3CLpro) | 16.47% | 18.88% | 9.00% | FORMIC ACID; (4S)-2-METHYL-2,4-PENTANEDIOL; SODIUM ION; tert-butyl-N-[(2S)-1-[(2S,4S)-4-methoxy-2-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]carbamoyl]pyrrolidin-1-yl]-3-methyl-1-oxidanylidene-butan-2-yl]carbamate | SODIUM ION; tert-butyl-N-[(2S)-1-[(2S,4S)-4-methoxy-2-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]carbamoyl]pyrrolidin-1-yl]-3-methyl-1-oxidanylidene-butan-2-yl]carbamate; FORMIC ACID; (4S)-2-METHYL-2,4-PENTANEDIOL | XDS | 99.90% | 15.0 | ELETTRA (11.2C) | PHENIX; PHENIX | 2023-03-28 | 37043904 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 88.4 | - | - | - | |||||||
| 8okm | 1.66 Å | 2023-05-03 | X-ray | MPD, NA, 84C, BR | SARS-CoV-2 | NSP5 (3CLpro) | 16.33% | 18.67% | 9.30% | SODIUM ION; tert-butyl-N-[(2S)-1-[(3S,3aS,6aR)-3-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]carbamoyl]-3,3a,4,5,6,6a-hexahydro-1H-cyclopenta[c]pyrrol-2-yl]-3-methyl-1-oxidanylidene-butan-2-yl]carbamate; (4S)-2-METHYL-2,4-PENTANEDIOL; BROMIDE ION | (4S)-2-METHYL-2,4-PENTANEDIOL; SODIUM ION; tert-butyl-N-[(2S)-1-[(3S,3aS,6aR)-3-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]carbamoyl]-3,3a,4,5,6,6a-hexahydro-1H-cyclopenta[c]pyrrol-2-yl]-3-methyl-1-oxidanylidene-butan-2-yl]carbamate; BROMIDE ION | XDS | 100.00% | 16.6 | ELETTRA (11.2C) | PHENIX; PHENIX | 2023-03-28 | 37043904 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 89.4 | - | - | - | |||||||
| 8okn | 1.35 Å | 2023-05-03 | X-ray | NA, MPD, 83W | SARS-CoV-2 | NSP5 (3CLpro) | 15.92% | 17.43% | 7.00% | tert-butyl-N-[(2S,3R)-3-[(2-methylpropan-2-yl)oxy]-1-oxidanylidene-1-[(2S)-2-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]carbamoyl]pyrrolidin-1-yl]butan-2-yl]carbamate; (4S)-2-METHYL-2,4-PENTANEDIOL; SODIUM ION | SODIUM ION; (4S)-2-METHYL-2,4-PENTANEDIOL; tert-butyl-N-[(2S,3R)-3-[(2-methylpropan-2-yl)oxy]-1-oxidanylidene-1-[(2S)-2-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]carbamoyl]pyrrolidin-1-yl]butan-2-yl]carbamate | XDS | 100.00% | 15.0 | ELETTRA (11.2C) | PHENIX; PHENIX | 2023-03-28 | 37043904 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 94.0 | - | - | - | |||||||
| 8oto | 1.80 Å | 2023-05-03 | X-ray | MES, AMP | SARS-CoV-2 | NSP10/NSP16 | 17.54% | 18.57% | 3.35% | ADENOSINE MONOPHOSPHATE; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID; ADENOSINE MONOPHOSPHATE | XDS | 99.82% | 12.65 | PETRA III, EMBL c/o DESY (P13 (MX1)) | PHENIX | 2023-04-21 | - | ZN | - | - | - | N/A | - | - | 2'-O-methyltransferase nsp16; Non-structural protein 10 | - | - | No | - | - | 89.9 | - | - | - | ||||||
| 7uw3 | 1.70 Å | 2023-05-10 | X-ray | - | SARS-CoV-2 | Nucleocapsid | 28.27% | 29.93% | 9.70% | - | XDS | 99.90% | 12.8 | SSRL (BL12-1) | PHENIX; PHENIX | 2022-05-02 | 35638584 | - | - | - | - | N/A | - | - | Nucleoprotein - N-terminal domain (UNP residues 40-174) | - | - | No | - | 7.4 | - | - | - | ||||||||
| 7xsa | 2.20 Å | 2023-05-10 | X-ray | - | SARS-CoV-2 | Spike | 23.12% | 28.31% | 9.80% | - | HKL-2000 | 99.10% | 5.5 | SSRF (BL18U1) | PHENIX | 2022-05-13 | 36914890 | - | - | - | - | N/A | - | - | P2S-2E9 Light chain; P2S-2E9 Heavy chain; Spike protein S1 - receptor binding domain | - | - | No | - | 12.6 | - | - | - | ||||||||
| 7xsb | 3.20 Å | 2023-05-10 | X-ray | - | SARS-CoV-2 | Spike | 25.76% | 32.43% | 17.10% | - | HKL-2000 | 98.50% | 2.8 | SSRF (BL18U1) | PHENIX | 2022-05-13 | 36914890 | - | - | - | - | N/A | - | - | P5S-3B11 Heavy chain; P5S-3B11 Light chain; Spike protein S1 - receptor binding domain | - | - | No | - | 4.7 | - | - | - | ||||||||
| 8a99 | 2.50 Å | 2023-05-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-06-28 | 36574772 | - | - | - | - | N/A | - | - | Fab47 (Light chain Variable domain); SARS-CoV2 spike 1-up conformation in complex with Fab47; Fab47 (Heavy chain variable domain) | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8fdw | 2.90 Å | 2023-05-10 | Cryo-EM | MAN, NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | alpha-D-mannopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | alpha-D-mannopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-12-05 | 37285872 | - | - | - | - | N/A | - | - | Spike protein S2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8h3m | 2.48 Å | 2023-05-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-10-09 | 37191569 | - | - | - | - | N/A | - | - | Spike glycoprotein; MO1 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8h3n | 2.73 Å | 2023-05-10 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-10-09 | 37191569 | - | - | - | - | N/A | - | - | MO1 heavy-chain; Spike glycoprotein; MO1 light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8ot0 | 2.21 Å | 2023-05-10 | X-ray | MTA, MES | SARS-CoV-2 | NSP10/NSP16 | 18.29% | 20.99% | 7.64% | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID; 5'-DEOXY-5'-METHYLTHIOADENOSINE | 5'-DEOXY-5'-METHYLTHIOADENOSINE; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | XDS | 99.86% | 7.36 | PETRA III, EMBL c/o DESY (P13 (MX1)) | PHENIX | 2023-04-20 | - | ZN | - | - | - | N/A | - | - | Non-structural protein 10; 2'-O-methyltransferase nsp16 | - | - | No | - | - | 74.2 | - | - | - | ||||||
| 8ov1 | 1.67 Å | 2023-05-10 | X-ray | MES, ADP, SAM | SARS-CoV-2 | NSP10/NSP16 | 17.38% | 18.78% | 3.49% | S-ADENOSYLMETHIONINE; ADENOSINE-5'-DIPHOSPHATE; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID; ADENOSINE-5'-DIPHOSPHATE; S-ADENOSYLMETHIONINE | XDS | 99.87% | 12.57 | PETRA III, EMBL c/o DESY (P13 (MX1)) | PHENIX | 2023-04-25 | - | ZN | - | - | - | N/A | - | - | Non-structural protein 10; 2'-O-methyltransferase nsp16 | - | - | No | - | - | 88.9 | - | - | - | ||||||
| 8ov2 | 1.86 Å | 2023-05-10 | X-ray | SGV, MES | SARS-CoV-2 | NSP10/NSP16 | 17.73% | 19.76% | 12.14% | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID; SANGIVAMYCIN | SANGIVAMYCIN; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | XDS | 99.94% | 9.75 | PETRA III, DESY (P11) | PHENIX | 2023-04-25 | - | ZN | - | - | - | N/A | - | - | Non-structural protein 10; 2'-O-methyltransferase nsp16 | - | - | No | - | - | 83.3 | - | - | - | ||||||
| 8ov3 | 1.82 Å | 2023-05-10 | X-ray | MES, 5ID | SARS-CoV-2 | NSP10/NSP16 | 18.20% | 21.06% | 16.79% | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID; (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL | XDS | 98.44% | 7.33 | PETRA III, DESY (P11) | PHENIX | 2023-04-25 | - | ZN | - | - | - | N/A | - | - | Non-structural protein 10; 2'-O-methyltransferase nsp16 | - | - | No | - | - | 73.6 | - | - | - | ||||||
| 8ov4 | 1.93 Å | 2023-05-10 | X-ray | MES, TO1 | SARS-CoV-2 | NSP10/NSP16 | 17.90% | 20.75% | 13.71% | 4-amino-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID; 4-amino-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile | XDS | 99.90% | 9.1 | PETRA III, DESY (P11) | PHENIX | 2023-04-25 | - | ZN | - | - | - | N/A | - | - | 2'-O-methyltransferase nsp16; Non-structural protein 10 | - | - | No | - | - | 76.2 | - | - | - | ||||||
| 8bse | 1.90 Å | 2023-05-17 | X-ray | TRS, NAG | SARS-CoV-2 | Spike | 17.69% | 21.61% | 10.90% | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL; 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 100.00% | 14.8 | MAX IV (BioMAX) | REFMAC | 2022-11-25 | 37141227 | - | - | - | - | N/A | - | - | 1D1 FAB LIGHT CHAIN; Spike glycoprotein; 1D1 FAB HEAVY CHAIN | - | - | No | - | 68.8 | - | - | - | |||||||
| 8bsf | 2.20 Å | 2023-05-17 | X-ray | NAG | SARS-CoV-2 | Spike | 19.59% | 22.94% | 9.60% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 98.90% | 13.0 | MAX IV (BioMAX) | REFMAC | 2022-11-25 | 37141227 | - | - | - | - | N/A | - | - | Spike protein S1; 3D2 FAB LIGHT CHAIN; 3D2 FAB HEAVY CHAIN | - | - | No | - | 55.6 | - | - | - | |||||||
| 8csj | 3.53 Å | 2023-05-17 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-05-12 | - | - | - | - | - | N/A | - | - | 4-33 heavy chain; 4-33 light chain; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 8dk8 | 2.60 Å | 2023-05-17 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 19.64% | 28.36% | N/A | - | XDS | 93.92% | 9.36 | CLSI (08B1-1) | PHENIX; PHENIX | 2022-07-04 | 37122453 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 12.5 | - | - | - | ||||||||
| 8dl9 | 1.90 Å | 2023-05-17 | X-ray | T4V | SARS-CoV-2 | NSP5 (3CLpro) | 18.10% | 22.04% | 6.50% | 1-{4-[(naphthalen-1-yl)methyl]piperazin-1-yl}ethan-1-one | 1-{4-[(naphthalen-1-yl)methyl]piperazin-1-yl}ethan-1-one | CrysalisPro | 99.90% | 15.17 | ROTATING ANODE () | PHENIX | 2022-07-07 | 36808989 | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | 64.7 | - | - | - | |||||||
| 8dlb | 1.90 Å | 2023-05-17 | X-ray | SRU | SARS-CoV-2 | NSP5 (3CLpro) | 17.24% | 20.61% | 10.10% | 1-[(5S)-5-(3,4-dimethoxyphenyl)-3-phenyl-4,5-dihydro-1H-pyrazol-1-yl]ethan-1-one | 1-[(5S)-5-(3,4-dimethoxyphenyl)-3-phenyl-4,5-dihydro-1H-pyrazol-1-yl]ethan-1-one | CrysalisPro | 99.70% | 14.42 | ROTATING ANODE () | PHENIX | 2022-07-07 | 36808989 | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | 77.2 | - | - | - | |||||||
| 8dmd | 2.00 Å | 2023-05-17 | X-ray | SVL | SARS-CoV-2 | NSP5 (3CLpro) | 20.96% | 24.02% | 7.00% | 1-[(3R)-4-[(3-chlorophenyl)methyl]-3-(2-methylpropyl)piperazin-1-yl]ethan-1-one | 1-[(3R)-4-[(3-chlorophenyl)methyl]-3-(2-methylpropyl)piperazin-1-yl]ethan-1-one | CrysalisPro | 99.80% | 14.02 | ROTATING ANODE () | PHENIX | 2022-07-08 | 36808989 | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | 44.6 | - | - | - | |||||||
| 8fwn | 1.50 Å | 2023-05-17 | X-ray | PO4 | SARS-CoV-2 | NSP3: PLpro | 18.16% | 19.83% | N/A | PHOSPHATE ION | PHOSPHATE ION | autoPROC | 99.99% | 17.26 | LNLS SIRUS (MANACA) | PHENIX | 2023-01-23 | - | ZN | - | - | - | N/A | - | - | Papain-like protease nsp3 | - | - | No | - | - | 82.8 | - | - | - | ||||||
| 8gia | 1.86 Å | 2023-05-17 | X-ray | ZJ3 | SARS-CoV-2 | NSP3: Macro | 20.84% | 25.82% | 18.40% | [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl [(2R,3S,4R,5R)-3,4-dihydroxy-5-{[2-oxo-4-(trifluoromethyl)-2H-1-benzopyran-7-yl]oxy}oxolan-2-yl]methyl dihydrogen diphosphate | [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl [(2R,3S,4R,5R)-3,4-dihydroxy-5-{[2-oxo-4-(trifluoromethyl)-2H-1-benzopyran-7-yl]oxy}oxolan-2-yl]methyl dihydrogen diphosphate | XDS | 99.50% | 6.9 | APS (24-ID-E) | PHENIX | 2023-03-13 | 37126856 | - | - | - | - | N/A | - | - | Non-structural protein 3 | - | - | No | - | 28.0 | - | - | - | |||||||
| 8hn6 | 2.07 Å | 2023-05-17 | X-ray | - | SARS-CoV-2 | Spike | 19.20% | 21.43% | N/A | - | xia2 | 99.90% | 18.7 | SSRF (BL19U1) | PHENIX | 2022-12-07 | 37234155 | - | - | - | - | N/A | - | - | Spike protein S1; Light chain of monoclonal antibody 3G10; Heavy chain of monoclonal antibody 3G10 | - | - | No | - | 70.4 | - | - | - | ||||||||
| 8hn7 | 3.00 Å | 2023-05-17 | X-ray | NAG | SARS-CoV-2 | Spike | 23.24% | 26.28% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 98.00% | 13.2 | SSRF (BL19U1) | PHENIX | 2022-12-07 | 37234155 | - | - | - | - | N/A | - | - | Spike protein S1; Heavy chain of monoclonal antibody 3C11; Light chain of monoclonal antibody 3C11 | - | - | No | - | 24.7 | - | - | - | |||||||
| 8if2 | 2.78 Å | 2023-05-17 | X-ray | NAG | SARS-CoV-2 | Spike | 19.96% | 24.47% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.70% | 14.4 | SPring-8 (BL32XU) | PHENIX | 2023-02-17 | 37169744 | ZN | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike protein S1 | - | - | No | - | 40.5 | - | - | - | |||||||
| 7xsc | 2.88 Å | 2023-05-24 | X-ray | - | SARS-CoV-2 | Spike | 20.04% | 24.97% | 20.30% | - | HKL-2000 | 99.40% | 4.4 | SSRF (BL18U1) | PHENIX | 2022-05-13 | 36914890 | - | - | - | - | N/A | - | - | P5S-2B10 Light chain; Spike protein S1 - receptor binding domain; P5S-2B10 Heavy chain | - | - | No | - | 35.6 | - | - | - | ||||||||
| 8cwi | 1.87 Å | 2023-05-24 | X-ray | - | SARS-CoV-2 | Spike | 16.99% | 20.49% | 7.30% | - | XDS | 98.50% | 14.5 | Australian Synchrotron (MX2) | PHENIX | 2022-05-19 | - | MG | - | - | - | N/A | - | - | Light chain of Fab arm of antibody 10G4; Spike protein S1 - receptor binding domain (RBD); Heavy chain of Fab arm of antibody 10G4 | - | - | No | - | - | 78.3 | - | - | - | |||||||
| 8cwj | 2.45 Å | 2023-05-24 | X-ray | NAG | Pangolin CoV | Spike | 19.62% | 23.01% | 11.00% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 100.00% | 11.8 | Australian Synchrotron (MX2) | PHENIX | 2022-05-19 | - | - | - | - | - | N/A | - | - | Light chain of CR3022 antibody Fab; Heavy chain of 4C12-B12 antibody Fab; Light chain of 4C12-B12 antibody Fab; Heavy chain of CR3022 antibody Fab; Spike glycoprotein | - | - | No | - | - | 54.8 | - | - | - | ||||||
| 8cwk | 2.37 Å | 2023-05-24 | X-ray | NAG | SARS-CoV-2 | Spike | 18.63% | 22.32% | 9.20% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.30% | 10.9 | Australian Synchrotron (MX2) | PHENIX | 2022-05-19 | - | - | - | - | - | N/A | - | - | Spike protein S1 - receptor binding domain (RBD); Light chain of Fab arm of antibody 4G1-C2; Heavy chain of Fab arm of antibody 4G1-C2; Light chain of Fab arm of antibody 10G4; Heavy chain of Fab arm of antibody 10G4 | - | - | No | - | - | 61.9 | - | - | - | ||||||
| 8dnn | 3.12 Å | 2023-05-24 | X-ray | NAG | SARS-CoV-2 | Spike | 19.25% | 25.20% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XPREP | 99.79% | 6.03 | APS (23-ID-B) | PHENIX | 2022-07-11 | - | - | - | - | - | N/A | - | - | Spike protein S1; 80 FAB LIGHT CHAIN; 80 FAB HEAVY CHAIN | - | - | No | - | - | 33.3 | - | - | - | ||||||
| 8gb5 | 3.35 Å | 2023-05-24 | X-ray | BCN | SARS-CoV-2 | Spike | 27.48% | 31.96% | 86.70% | BICINE | BICINE | HKL-2000 | 91.50% | 1.3 | APS (23-ID-D) | PHENIX | 2023-02-24 | 37163615 | - | - | - | - | N/A | - | - | 25F9 Heavy chain; Spike protein S1 - Receptor binding domain, UNP residues 333-530; 25F9 Light chain | - | - | No | - | 5.0 | - | - | - | |||||||
| 8gb6 | 1.75 Å | 2023-05-24 | X-ray | - | SARS-CoV-2 | Spike | 18.21% | 22.07% | 99.10% | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | - | HKL-2000 | 97.20% | 4.7 | APS (23-ID-B) | PHENIX | 2023-02-24 | 37163615 | - | - | - | - | N/A | - | - | 21B6 Heavy chain; 21B6 Light chain; Spike protein S1 - Receptor binding domain, UNP residues 333-530 | - | - | No | - | 64.5 | - | - | - | |||||||
| 8gb7 | 2.57 Å | 2023-05-24 | X-ray | - | SARS-CoV-2 | Spike | 20.39% | 24.48% | 98.60% | - | HKL-2000 | 98.90% | 4.2 | APS (23-ID-B) | PHENIX | 2023-02-24 | 37163615 | - | - | - | - | N/A | - | - | 20A7 Heavy chain; 20A7 Light chain; Spike protein S1 - Receptor binding domain, UNP residues 333-530 | - | - | No | - | 40.4 | - | - | - | ||||||||
| 8gb8 | 2.30 Å | 2023-05-24 | X-ray | NAG | SARS-CoV-2 | Spike | 19.95% | 22.83% | 99.00% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.40% | 5.8 | SSRL (BL12-1) | PHENIX | 2023-02-24 | 37163615 | - | - | - | - | N/A | - | - | Spike protein S1 - Receptor binding domain, UNP residues 319-541; 20A7 Light chain; 20A7 Heavy chain | - | - | No | - | 56.7 | - | - | - | |||||||
| 8ilc | 2.20 Å | 2023-05-24 | X-ray | - | SARS-CoV-2 | NSP3(C-term) | 21.05% | 23.92% | N/A | - | XDS | 99.90% | 19.3 | PETRA III, DESY (P11) | PHENIX | 2023-03-03 | - | - | - | - | - | N/A | - | - | Papain-like protease nsp3 - CoV-Y domain | - | - | No | - | - | 45.6 | - | - | - | |||||||
| 8ios | 2.50 Å | 2023-05-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2023-03-13 | 37193706 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8iot | 2.51 Å | 2023-05-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2023-03-13 | 37193706 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8iou | 3.18 Å | 2023-05-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2023-03-13 | 37193706 | - | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8iov | 3.29 Å | 2023-05-24 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2023-03-13 | 37193706 | - | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8otr | 1.77 Å | 2023-05-24 | X-ray | SAM, W08, MES | SARS-CoV-2 | NSP10/NSP16 | 18.80% | 20.95% | 5.61% | (2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-N-(1-methylpiperidin-4-yl)-3,4-bis(oxidanyl)oxolane-2-carboxamide; S-ADENOSYLMETHIONINE; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | S-ADENOSYLMETHIONINE; (2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-N-(1-methylpiperidin-4-yl)-3,4-bis(oxidanyl)oxolane-2-carboxamide; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | XDS | 99.79% | 8.41 | PETRA III, DESY (P11) | PHENIX | 2023-04-21 | - | ZN | - | - | - | N/A | - | - | Non-structural protein 10; 2'-O-methyltransferase nsp16 | - | - | No | - | - | 74.5 | - | - | - | ||||||
| 8spj | 2.08 Å | 2023-05-24 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 21.68% | 24.18% | 10.40% | - | HKL-2000 | 98.50% | 22.15 | APS (19-ID) | REFMAC | 2023-05-03 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 43.3 | - | - | - | |||||||
| 7vzt | 3.41 Å | 2023-05-31 | X-ray | - | SARS-CoV-2 | Spike | 21.75% | 26.26% | N/A | - | XDS | 71.90% | 12.6 | SSRF (BL19U1) | PHENIX | 2021-11-16 | - | - | - | - | - | N/A | - | - | Spike protein S1; GH12-Heavy; GH12-LIGHT | - | - | No | - | - | 24.7 | - | - | - | |||||||
| 7xeg | 2.69 Å | 2023-05-31 | X-ray | - | SARS-CoV-2 | Spike | 19.50% | 24.76% | 18.90% | - | HKL-2000 | 99.50% | 10.0 | SSRF (BL19U1) | PHENIX | 2022-03-31 | - | - | - | - | - | N/A | - | - | CB6-092-Fab heavy chain; Spike protein S1; CB6-092-Fab light chain | - | - | No | - | - | 37.6 | - | - | - | |||||||
| 7xei | 2.76 Å | 2023-05-31 | X-ray | - | SARS-CoV-2 | Spike | 21.53% | 26.29% | 7.40% | - | XDS | 91.80% | 11.1 | SSRF (BL10U2) | PHENIX | 2022-03-31 | - | - | - | - | - | N/A | - | - | CB6-092-Fab light chain; CB6-092-Fab heavy chain; Spike protein S1 | - | - | No | - | - | 24.6 | - | - | - | |||||||
| 7xjw | 2.75 Å | 2023-05-31 | X-ray | K36 | CCoV | NSP5 (3CLpro) | 21.25% | 23.28% | 6.20% | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | HKL-2000 | 99.30% | 20.39 | NSRRC (TPS 05A) | REFMAC | 2022-04-18 | 35628479 | - | - | - | - | N/A | - | - | ORF1a polyprotein | - | - | No | - | 52.4 | - | - | - | |||||||
| 7xmn | 2.30 Å | 2023-05-31 | X-ray | NAG, NNH, MES | SARS-CoV-2 | ORF8 | 21.49% | 24.53% | N/A | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID; NOR-N-OMEGA-HYDROXY-L-ARGININE; 2-acetamido-2-deoxy-beta-D-glucopyranose; alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose; NOR-N-OMEGA-HYDROXY-L-ARGININE; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | XDS | 96.80% | 7.9 | SSRF (BL18U1) | PHENIX; PHENIX | 2022-04-26 | - | - | - | - | - | N/A | - | - | Maltodextrin-binding protein; ORF8 protein | - | - | No | - | - | 39.8 | - | - | - | ||||||
| 8h07 | 2.90 Å | 2023-05-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-09-28 | 37069158 | - | - | - | - | N/A | - | - | 10-5B L chain; 6-2C L chain; 6-2C H chain; 10-5B H chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8h08 | 3.20 Å | 2023-05-31 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-09-28 | 37069158 | - | - | - | - | N/A | - | - | Spike glycoprotein; 10-5B H chain; 6-2C L chain; 6-2C H chain; 10-5B L chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8osx | 1.83 Å | 2023-05-31 | X-ray | SAM, MES, ATP | SARS-CoV-2 | NSP10/NSP16 | 19.74% | 22.32% | 19.51% | S-ADENOSYLMETHIONINE; ADENOSINE-5'-TRIPHOSPHATE; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | S-ADENOSYLMETHIONINE; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID; ADENOSINE-5'-TRIPHOSPHATE | XDS | 99.55% | 6.85 | PETRA III, DESY (P11) | PHENIX | 2023-04-20 | - | ZN | - | - | - | N/A | - | - | 2'-O-methyltransferase nsp16; Non-structural protein 10 | - | - | No | - | - | 61.9 | - | - | - | ||||||
| 7wt7 | 3.40 Å | 2023-06-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-04 | 35977939 | - | - | - | - | N/A | - | - | Heavy chain of Fab 9A8; Spike glycoprotein; Light chain of Fab 9A8 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wt8 | 3.60 Å | 2023-06-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-02-04 | 35977939 | - | - | - | - | N/A | - | - | light chain of Fab 9A8; Heavy chain of Fab 9A8; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7wt9 | 4.30 Å | 2023-06-07 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-02-04 | 35977939 | - | - | - | - | N/A | - | - | Light chain of Fab 9A8; Heavy chain of Fab 9A8; Spike glycoprotein - RBD | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7y6d | 4.39 Å | 2023-06-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-06-20 | 37094167 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 8d4p | 2.04 Å | 2023-06-07 | X-ray | QAO | SARS-CoV-2 | NSP5 (3CLpro) | 18.90% | 25.66% | 7.90% | 2-chloro-N-[(1R)-2-{[2-(3-fluorophenyl)ethyl]amino}-2-oxo-1-(pyridin-3-yl)ethyl]-N-[4-(pentafluoro-lambda~6~-sulfanyl)phenyl]acetamide | 2-chloro-N-[(1R)-2-{[2-(3-fluorophenyl)ethyl]amino}-2-oxo-1-(pyridin-3-yl)ethyl]-N-[4-(pentafluoro-lambda~6~-sulfanyl)phenyl]acetamide | HKL-3000 | 99.00% | 20.66 | APS (19-ID) | REFMAC | 2022-06-02 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 29.5 | - | - | - | ||||||
| 8d55 | 2.80 Å | 2023-06-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-06-04 | 37430064 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8d56 | 3.00 Å | 2023-06-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-06-04 | 37430064 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8d5a | 3.10 Å | 2023-06-07 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-06-04 | 37430064 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8sk4 | 2.00 Å | 2023-06-07 | X-ray | IMD, I3R | SARS-CoV-2 | NSP5 (3CLpro) | 19.36% | 23.59% | 7.80% | IMIDAZOLE; 2-chloro-1-[(5R)-3-phenyl-5-(quinoxalin-5-yl)-4,5-dihydro-1H-pyrazol-1-yl]ethan-1-one | IMIDAZOLE; 2-chloro-1-[(5R)-3-phenyl-5-(quinoxalin-5-yl)-4,5-dihydro-1H-pyrazol-1-yl]ethan-1-one | XDS | 92.20% | 15.5 | APS (17-ID) | BUSTER | 2023-04-18 | 37069799 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 49.1 | - | - | - | |||||||
| 8skh | 1.88 Å | 2023-06-07 | X-ray | A1W | SARS-CoV-2 | NSP5 (3CLpro) | 18.44% | 23.07% | 3.90% | 2-chloro-1-[(4R,5R)-3,4,5-triphenyl-4,5-dihydro-1H-pyrazol-1-yl]ethan-1-one | 2-chloro-1-[(4R,5R)-3,4,5-triphenyl-4,5-dihydro-1H-pyrazol-1-yl]ethan-1-one | XDS | 82.70% | 18.6 | APS (17-ID) | BUSTER | 2023-04-19 | 37069799 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 54.4 | - | - | - | |||||||
| 8a23 | 2.80 Å | 2023-06-14 | X-ray | KW6 | SARS-CoV-2 | NSP10/NSP16 | 23.30% | 25.58% | 44.75% | (2R,3R,4S,5R)-2-[4-azanyl-5-(2-quinolin-3-ylethynyl)pyrrolo[2,3-d]pyrimidin-7-yl]-5-(hydroxymethyl)oxolane-3,4-diol | (2R,3R,4S,5R)-2-[4-azanyl-5-(2-quinolin-3-ylethynyl)pyrrolo[2,3-d]pyrimidin-7-yl]-5-(hydroxymethyl)oxolane-3,4-diol | XDS | 99.73% | 6.07 | ROTATING ANODE () | PHENIX | 2022-06-02 | - | ZN | - | - | - | N/A | - | - | Non-structural protein 10; 2'-O-methyltransferase nsp16 | - | - | No | - | - | 30.2 | - | - | - | ||||||
| 8d34 | 2.91 Å | 2023-06-14 | X-ray | - | SARS-CoV-2 | NSP15 | 24.62% | 26.26% | 26.20% | - | HKL-2000 | 100.00% | 4.3 | ROTATING ANODE () | PHENIX | 2022-05-31 | - | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease nsp15 | - | - | No | - | - | 24.7 | - | - | - | |||||||
| 8dt8 | 3.34 Å | 2023-06-14 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-07-25 | 37276407 | - | - | - | - | N/A | - | - | Nb136 nanobody; Spike glycoprotein; LM18 nanobody | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8eoo | 2.77 Å | 2023-06-14 | X-ray | NAG | SARS-CoV-2 | Spike | 19.37% | 25.51% | 14.00% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 98.70% | 11.0 | APS (24-ID-E) | PHENIX | 2022-10-03 | 37395646 | - | - | - | - | N/A | - | - | WRAIR-2063 antibody Fab heavy chain; Spike protein S1 - receptor binding domain (RBD); WRAIR-2151 antibody Fab heavy chain; WRAIR-2063 antibody Fab light chain; WRAIR-2151 antibody Fab light chain | - | - | No | - | 30.6 | - | - | - | |||||||
| 8sxo | 2.76 Å | 2023-06-14 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 20.14% | 24.36% | 9.60% | - | HKL-2000 | 93.00% | 12.21 | APS (19-ID) | REFMAC | 2023-05-23 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 41.5 | - | - | - | |||||||
| 7y6l | 3.50 Å | 2023-06-21 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-06-21 | 37288342 | - | - | - | - | N/A | - | - | Spike glycoprotein; Ab816 light chain; Ab816 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7y6n | 4.40 Å | 2023-06-21 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-06-21 | 37288342 | - | - | - | - | N/A | - | - | Spike glycoprotein; Ab803 light chain; Ab803 heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8epn | 2.80 Å | 2023-06-21 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-10-06 | 37285422 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8epp | 3.10 Å | 2023-06-21 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-10-06 | 37285422 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8epq | 3.30 Å | 2023-06-21 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-10-06 | 37285422 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8hur | 1.64 Å | 2023-06-21 | X-ray | 7YY | SARS-CoV-2 | NSP5 (3CLpro) | 20.07% | 22.61% | 2.80% | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | XDS | 93.20% | 19.2 | SSRF (BL02U1) | PHENIX; PHENIX | 2022-12-24 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 59.0 | - | - | - | ||||||
| 8hus | 1.97 Å | 2023-06-21 | X-ray | 7YY | SARS-CoV | NSP5 (3CLpro) | 20.55% | 23.72% | 2.60% | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | XDS | 95.20% | 10.1 | SSRF (BL02U1) | PHENIX; PHENIX | 2022-12-24 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 47.6 | - | - | - | ||||||
| 8hut | 1.98 Å | 2023-06-21 | X-ray | 7YY | MERS-CoV | NSP5 (3CLpro) | 21.39% | 25.38% | 12.30% | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | XDS | 97.10% | 14.9 | SSRF (BL02U1) | PHENIX; PHENIX | 2022-12-24 | - | - | - | - | - | N/A | - | - | ORF1a | - | - | No | - | - | 31.9 | - | - | - | ||||||
| 8huu | 1.71 Å | 2023-06-21 | X-ray | 7YY | HCoV-NL63 | NSP5 (3CLpro) | 18.82% | 21.73% | 4.60% | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | XDS | 98.10% | 18.8 | SSRF (BL02U1) | PHENIX; PHENIX | 2022-12-24 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 67.7 | - | - | - | ||||||
| 8huv | 1.97 Å | 2023-06-21 | X-ray | 7YY | SARS-CoV-2 | NSP5 (3CLpro) | 20.74% | 24.71% | 4.90% | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | XDS | 98.20% | 11.6 | SSRF (BL02U1) | PHENIX; PHENIX | 2022-12-24 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 38.0 | - | - | - | ||||||
| 8huw | 1.75 Å | 2023-06-21 | X-ray | 7YY | SARS-CoV-2 | NSP5 (3CLpro) | 19.63% | 22.79% | 4.10% | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | XDS | 97.70% | 18.7 | SSRF (BL02U1) | PHENIX; PHENIX | 2022-12-24 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 57.3 | - | - | - | ||||||
| 8hux | 1.74 Å | 2023-06-21 | X-ray | 7YY | SARS-CoV-2 | NSP5 (3CLpro) | 20.67% | 24.49% | 3.80% | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | XDS | 99.10% | 27.4 | SSRF (BL02U1) | PHENIX; PHENIX | 2022-12-24 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 40.4 | - | - | - | ||||||
| 8idn | 3.35 Å | 2023-06-21 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2023-02-13 | 37428848 | - | - | - | - | N/A | - | - | E77 Fab heavy chain; E77 Fab light chain; Spike protein S1 - Receptor binding domain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8j1q | 3.30 Å | 2023-06-21 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; AM047-0; AM032-0 | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2023-04-13 | 37375202 | - | - | - | - | N/A | - | - | AM047-0; Fab Light chain (REGN10987); Fab heavy chain (REGN10987); AM032-0; Spike protein S1 - RBD | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8j26 | 3.40 Å | 2023-06-21 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | AM047-6; 2-acetamido-2-deoxy-beta-D-glucopyranose; AM032-4 | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2023-04-14 | 37375202 | - | - | - | - | N/A | - | - | Fab light chain (REGN10987); AM047-6; Spike protein S1 - RBD; Fab heavy chain (REGN10987); AM032-4 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7ytn | 3.51 Å | 2023-06-28 | X-ray | - | SARS-CoV-2 | Spike | 23.67% | 27.35% | N/A | - | HKL-2000 | 91.80% | 9.8 | PAL/PLS (5C (4A)) | PHENIX | 2022-08-15 | 36466915 | - | - | - | - | N/A | - | - | Spike protein S1 - RBD; Heavy chain of Fab; Light chain of Fab | - | - | No | - | 17.5 | - | - | - | ||||||||
| 7yve | 3.40 Å | 2023-06-28 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-08-19 | 37322000 | - | - | - | - | N/A | - | - | Spike glycoprotein; TH027 Fab light chain; TH027 Fab heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7yvf | 3.40 Å | 2023-06-28 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-08-19 | 37322000 | - | - | - | - | N/A | - | - | TH027 Fab light chain; Spike glycoprotein; TH027 Fab heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7yvg | 3.40 Å | 2023-06-28 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-08-19 | 37322000 | - | - | - | - | N/A | - | - | TH132 Fab heavy chain; TH132 Fab light chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7yvh | 3.60 Å | 2023-06-28 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-08-19 | 37322000 | - | - | - | - | N/A | - | - | Spike glycoprotein; TH132 Fab heavy chain; TH132 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7yvi | 3.70 Å | 2023-06-28 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-08-19 | 37322000 | - | - | - | - | N/A | - | - | Spike glycoprotein; TH236 Fab heavy chain; TH236 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7yvj | 3.60 Å | 2023-06-28 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-08-19 | 37322000 | - | - | - | - | N/A | - | - | Spike glycoprotein; TH236 Fab heavy chain; TH236 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7yvk | 3.20 Å | 2023-06-28 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-08-19 | 37322000 | - | - | - | - | N/A | - | - | Spike glycoprotein; TH272 Fab heavy chain; TH272 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7yvl | 3.30 Å | 2023-06-28 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-08-19 | 37322000 | - | - | - | - | N/A | - | - | Spike glycoprotein; TH272 Fab light chain; TH272 Fab heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7yvm | 3.50 Å | 2023-06-28 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-08-19 | 37322000 | - | - | - | - | N/A | - | - | TH281 Fab heavy chain; TH281 Fab light chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7yvn | 3.40 Å | 2023-06-28 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-08-19 | 37322000 | - | - | - | - | N/A | - | - | TH281 Fab heavy chain; Spike glycoprotein; TH281 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7yvo | 3.30 Å | 2023-06-28 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-08-19 | 37322000 | - | - | - | - | N/A | - | - | TH27 Fab light chain; TH132 Fab heavy chain; TH27 Fab heavy chain; TH132 Fab light chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7yvp | 3.80 Å | 2023-06-28 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-08-19 | 37322000 | - | - | - | - | N/A | - | - | Spike glycoprotein; TH272 Fab heavy chain; TH272 Fab light chain; TH281 Fab light chain; TH281 Fab heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7z8o | 0.96 Å | 2023-06-28 | X-ray | KZ0 | SARS-CoV-2 | Spike | 15.20% | 16.91% | 6.10% | Stapled peptide; 2,4,6-tris(chloromethyl)-1,3,5-triazine | 2,4,6-tris(chloromethyl)-1,3,5-triazine | XDS | 99.20% | 19.6 | Diamond (I04) | REFMAC | 2022-03-18 | 37328472 | - | - | - | - | N/A | - | - | Stapled peptide; Spike protein S1 | - | - | No | - | 95.3 | - | - | - | |||||||
| 8aaa | 1.90 Å | 2023-06-28 | X-ray | 29N | SARS-CoV-2 | Spike | 22.67% | 23.57% | 24.70% | 1,1',1''-(1,3,5-triazinane-1,3,5-triyl)tripropan-1-one; Stapled peptide | 1,1',1''-(1,3,5-triazinane-1,3,5-triyl)tripropan-1-one | XDS | 91.50% | 9.3 | Diamond (I04) | BUSTER | 2022-06-30 | 37328472 | - | - | - | - | N/A | - | - | Stapled peptide; Spike protein S1 | - | - | No | - | 49.3 | - | - | - | |||||||
| 8bon | 3.20 Å | 2023-06-28 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; Macrocyclic peptide S1B3inL1 | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-11-15 | 37339194 | - | - | - | - | N/A | - | - | Macrocyclic peptide S1B3inL1; Spike glycoprotein,Fibritin | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8ddl | 1.95 Å | 2023-06-28 | X-ray | NA, PGE | SARS-CoV-2 | NSP5 (3CLpro) | 16.83% | 20.67% | 16.90% | TRIETHYLENE GLYCOL | SODIUM ION; TRIETHYLENE GLYCOL | DIALS | 100.00% | 6.4 | CHESS (F1) | PHENIX | 2022-06-18 | 37699927 | - | - | - | - | N/A | - | - | ORF1a polyprotein; ORF1a polyprotein | - | - | No | - | 76.8 | - | - | - | |||||||
| 8dib | 2.17 Å | 2023-06-28 | X-ray | TKX | SARS-CoV-2 | NSP5 (3CLpro) | 22.12% | 27.64% | 16.30% | 5-bromo-3-[(4-chloro-3-nitrophenyl)methoxy]pyridine-2-carbaldehyde | 5-bromo-3-[(4-chloro-3-nitrophenyl)methoxy]pyridine-2-carbaldehyde | XDS | 98.60% | 8.5 | SSRL (BL12-2) | PHENIX | 2022-06-29 | 37354015 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 15.9 | - | - | - | |||||||
| 8dic | 2.09 Å | 2023-06-28 | X-ray | TNI | SARS-CoV-2 | NSP5 (3CLpro) | 22.51% | 28.17% | 15.00% | 5-bromo-3-[(3-bromo-4-chlorophenyl)methoxy]pyridine-2-carbaldehyde | 5-bromo-3-[(3-bromo-4-chlorophenyl)methoxy]pyridine-2-carbaldehyde | XDS | 98.50% | 8.8 | SSRL (BL12-2) | PHENIX | 2022-06-29 | 37354015 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 13.4 | - | - | - | |||||||
| 8die | 1.90 Å | 2023-06-28 | X-ray | U1J | SARS-CoV-2 | NSP5 (3CLpro) | 25.37% | 30.35% | 46.00% | 5-bromo-3-[(4-methyl-3-nitrophenyl)methoxy]pyridine-2-carbaldehyde | 5-bromo-3-[(4-methyl-3-nitrophenyl)methoxy]pyridine-2-carbaldehyde | XDS | 98.50% | 4.5 | SSRL (BL12-2) | PHENIX | 2022-06-29 | 37354015 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 6.8 | - | - | - | |||||||
| 8dif | 1.98 Å | 2023-06-28 | X-ray | U1R | SARS-CoV-2 | NSP5 (3CLpro) | 24.03% | 29.44% | 9.20% | 5-bromo-3-[(naphthalen-2-yl)methoxy]pyridine-2-carbaldehyde | 5-bromo-3-[(naphthalen-2-yl)methoxy]pyridine-2-carbaldehyde | XDS | 96.80% | 12.3 | ALS (8.3.1) | PHENIX | 2022-06-29 | 37354015 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 8.7 | - | - | - | |||||||
| 8dig | 2.45 Å | 2023-06-28 | X-ray | U26 | SARS-CoV-2 | NSP5 (3CLpro) | 21.42% | 27.05% | 32.40% | (3P)-1-[(4-fluorophenyl)methyl]-3-(isoquinolin-4-yl)imidazolidine-2,4-dione | (3P)-1-[(4-fluorophenyl)methyl]-3-(isoquinolin-4-yl)imidazolidine-2,4-dione | XDS | 97.90% | 10.3 | ALS (8.3.1) | PHENIX | 2022-06-29 | 37354015 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 19.2 | - | - | - | |||||||
| 8dih | 2.12 Å | 2023-06-28 | X-ray | U2B | SARS-CoV-2 | NSP5 (3CLpro) | 21.14% | 27.60% | 4.90% | (1P,1'R)-1-(isoquinolin-4-yl)-2',3'-dihydrospiro[imidazolidine-4,1'-indene]-2,5-dione | (1P,1'R)-1-(isoquinolin-4-yl)-2',3'-dihydrospiro[imidazolidine-4,1'-indene]-2,5-dione | XDS | 97.00% | 19.1 | ALS (8.3.1) | PHENIX | 2022-06-29 | 37354015 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 16.1 | - | - | - | |||||||
| 8dii | 2.59 Å | 2023-06-28 | X-ray | U2I | SARS-CoV-2 | NSP5 (3CLpro) | 21.45% | 27.13% | 18.20% | (2S)-N-(isoquinolin-4-yl)-2-methyl-2,3-dihydro-1,4-benzoxazepine-4(5H)-carboxamide | (2S)-N-(isoquinolin-4-yl)-2-methyl-2,3-dihydro-1,4-benzoxazepine-4(5H)-carboxamide | XDS | 99.90% | 8.6 | ALS (8.3.1) | PHENIX | 2022-06-29 | 37354015 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 18.8 | - | - | - | |||||||
| 8e1g | 2.57 Å | 2023-06-28 | X-ray | NAG | SARS-CoV-2 | Spike | 19.59% | 24.39% | 8.00% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.40% | 16.1 | APS (31-ID) | PHENIX | 2022-08-10 | 37083327 | - | - | - | - | N/A | - | - | 2A10 Fab, heavy chain; 2A10 Fab, light chain; Spike protein S1 | - | - | No | - | 41.3 | - | - | - | |||||||
| 8gou | 3.70 Å | 2023-06-28 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-08-25 | 37322000 | - | - | - | - | N/A | - | - | TH003 Fab heavy chain; TH003 Fab light chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8gpy | 2.51 Å | 2023-06-28 | X-ray | - | SARS-CoV-2 | Spike | 21.49% | 25.07% | 12.50% | - | HKL-2000 | 100.00% | 6.3 | SSRF (BL10U2) | PHENIX | 2022-08-27 | 37322000 | - | - | - | - | N/A | - | - | scFv light chain; Spike protein S1; scFv heavy chain | - | - | No | - | 34.6 | - | - | - | ||||||||
| 8smt | 3.16 Å | 2023-06-28 | X-ray | NAG | SARS-CoV-2 | Spike | 21.44% | 24.86% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.70% | 7.15 | APS (24-ID-E) | PHENIX | 2023-04-26 | - | - | - | - | - | N/A | - | - | WRAIR-2134 Fab light chain; WRAIR-2134 Fab heavy chain; Spike protein S1 - receptor binding domain | - | - | No | - | - | 36.6 | - | - | - | |||||||
| 7yc5 | 3.10 Å | 2023-07-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-06-30 | - | - | - | - | - | N/A | - | - | Heavy chain from K202.B, bispecific antibody; Spike glycoprotein; Light chain from K202.B, bispecific antibody | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7yco | 1.96 Å | 2023-07-05 | X-ray | NAG | SARS-CoV-2 | Spike | 23.55% | 25.85% | 8.47% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.93% | 19.88 | PAL/PLS (5C (4A)) | PHENIX | 2022-07-01 | - | - | - | - | - | N/A | - | - | Spike protein S1; Repebody (A6) | - | - | No | - | - | 27.8 | - | - | - | ||||||
| 8cim | 3.00 Å | 2023-07-05 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2023-02-10 | - | - | - | - | - | N/A | - | - | BA.2-07 FAB LIGHT CHAIN; BA.2-07 FAB HEAVY CHAIN; Spike glycoprotein,Fibritin | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 8did | 1.95 Å | 2023-07-05 | X-ray | U0R | SARS-CoV-2 | NSP5 (3CLpro) | 25.39% | 29.42% | 17.00% | 5-bromo-3-[(5-bromo-2-chlorophenyl)methoxy]pyridine-2-carbaldehyde | 5-bromo-3-[(5-bromo-2-chlorophenyl)methoxy]pyridine-2-carbaldehyde | XDS | 98.60% | 7.4 | SSRL (BL12-2) | PHENIX | 2022-06-29 | 37354015 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 8.7 | - | - | - | |||||||
| 8elo | 2.72 Å | 2023-07-05 | X-ray | NAG | SARS-CoV-2 | Spike | 22.00% | 26.12% | 11.70% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 96.90% | 8.4 | SSRL (BL12-1) | PHENIX | 2022-09-26 | 37276407 | - | - | - | - | N/A | - | - | CC12.1 Fab light chain; Spike protein S1 - Receptor binding domain, UNP residues 333-530; Nanobody Nb-C4-225; CC12.1 Fab heavy chain | - | - | No | - | 25.7 | - | - | - | |||||||
| 8elp | 2.83 Å | 2023-07-05 | X-ray | NAG | SARS-CoV-2 | Spike | 23.81% | 28.38% | 95.50% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 96.80% | 4.7 | SSRL (BL12-2) | PHENIX | 2022-09-26 | 37276407 | - | - | - | - | N/A | - | - | CC12.1 Fab light chain; CC12.1 Fab heavy chain; Nanobody Nb-C4-240; Spike protein S1 - Receptor binding domain, UNP residues 333-530 | - | - | No | - | 12.5 | - | - | - | |||||||
| 8elq | 2.21 Å | 2023-07-05 | X-ray | NAG | SARS-CoV-2 | Spike | 23.44% | 28.49% | 13.10% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 96.40% | 5.7 | APS (23-ID-B) | PHENIX | 2022-09-26 | 37276407 | - | - | - | - | N/A | - | - | CC12.1 Fab heavy chain; CC12.1 Fab light chain; Nanobody Nb-C4-255; Spike protein S1 - Receptor binding domain, UNP residues 333-530 | - | - | No | - | 12.0 | - | - | - | |||||||
| 7xd2 | 3.30 Å | 2023-07-12 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2022-03-26 | 37069158 | - | - | - | - | N/A | - | - | H chain of antibody 10-5B; Spike glycoprotein; L chian of antibody 10-5B | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 7y1y | 3.30 Å | 2023-07-12 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-06-09 | 37376697 | - | - | - | - | N/A | - | - | Spike glycoprotein; Processed angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7y1z | 3.40 Å | 2023-07-12 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-06-09 | 37376697 | - | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7ybi | 4.10 Å | 2023-07-12 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-06-29 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 7ybj | 3.73 Å | 2023-07-12 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2022-06-29 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||||
| 7yd1 | 2.99 Å | 2023-07-12 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-07-02 | - | - | - | - | - | N/A | - | - | Spike protein S1 - RBD; XG005-VL; XG005-VH | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 8gfk | 2.00 Å | 2023-07-12 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 16.85% | 20.61% | 9.00% | - | CrysalisPro | 99.90% | 13.2 | ROTATING ANODE () | PHENIX | 2023-03-08 | 37271339 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 77.2 | - | - | - | ||||||||
| 8gfn | 1.80 Å | 2023-07-12 | X-ray | ZGI | SARS-CoV-2 | NSP5 (3CLpro) | 17.19% | 20.62% | 5.90% | (1R,2S,5S)-N-{(2S)-1-(1,3-benzothiazol-2-yl)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(2S)-1-(1,3-benzothiazol-2-yl)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | CrysalisPro | 93.70% | 10.3 | ROTATING ANODE () | PHENIX | 2023-03-08 | 37271339 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 77.1 | - | - | - | |||||||
| 8gfo | 2.00 Å | 2023-07-12 | X-ray | ZH0 | SARS-CoV-2 | NSP5 (3CLpro) | 16.84% | 20.15% | 8.60% | N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | CrysalisPro | 99.60% | 11.2 | ROTATING ANODE () | PHENIX | 2023-03-08 | 37271339 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 80.7 | - | - | - | |||||||
| 8gfr | 2.00 Å | 2023-07-12 | X-ray | ZGO | SARS-CoV-2 | NSP5 (3CLpro) | 16.34% | 20.15% | 6.60% | (1R,2S,5S)-N-{(1S)-1-cyano-2-[(3S)-2-oxopyrrolidin-3-yl]ethyl}-6,6-dimethyl-3-[3-methyl-N-({1-[(2-methylpropane-2-sulfonyl)methyl]cyclohexyl}carbamoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1S)-1-cyano-2-[(3S)-2-oxopyrrolidin-3-yl]ethyl}-6,6-dimethyl-3-[3-methyl-N-({1-[(2-methylpropane-2-sulfonyl)methyl]cyclohexyl}carbamoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | CrysalisPro | 94.90% | 16.5 | ROTATING ANODE () | PHENIX | 2023-03-08 | 37271339 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 80.7 | - | - | - | |||||||
| 8gfu | 1.80 Å | 2023-07-12 | X-ray | ZGW | SARS-CoV-2 | NSP5 (3CLpro) | 16.56% | 19.33% | 4.80% | Nirmatrelvir | Nirmatrelvir | CrysalisPro | 99.30% | 20.3 | ROTATING ANODE () | PHENIX | 2023-03-08 | 37271339 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 86.0 | - | - | - | |||||||
| 8gqc | 1.35 Å | 2023-07-12 | X-ray | - | SARS-CoV-2 | NSP3(SUD-M) | 13.95% | 16.23% | 4.20% | - | XDS | 96.00% | 1.2 | SLS (X06DA) | PHENIX | 2022-08-30 | - | - | - | - | - | N/A | - | - | Papain-like protease nsp3 - SUD domain | - | - | No | - | - | 96.7 | - | - | - | |||||||
| 8gto | 3.20 Å | 2023-07-12 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-09-08 | 37015915 | - | - | - | - | N/A | - | - | heavy chain of XGv282; Spike glycoprotein; light chain of XGv282 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8gtp | 3.10 Å | 2023-07-12 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-09-08 | 37015915 | - | - | - | - | N/A | - | - | heavy chain of XGv289; light chain of XGv289; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8gtq | 3.10 Å | 2023-07-12 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-09-08 | 37015915 | - | - | - | - | N/A | - | - | light chain of S2L20; Spike glycoprotein; heavy chain of S2L20 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8hbl | 1.58 Å | 2023-07-12 | X-ray | PO4, LI | SARS-CoV-2 | NSP3(SUD-M) | 13.87% | 17.96% | 7.10% | PHOSPHATE ION; LITHIUM ION | PHOSPHATE ION; LITHIUM ION | XDS | 99.60% | 21.99 | SLS (X10SA) | PHENIX | 2022-10-29 | - | LI | - | - | - | N/A | - | - | Non-structural protein 3 | - | - | No | - | - | 92.3 | - | - | - | ||||||
| 7y6k | 3.34 Å | 2023-07-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-06-20 | - | - | - | - | - | N/A | - | - | Fab region of heavy chain from K202.B, bispecific antibody; scFv region of light chain from K202.B, bispecific antibody; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7ybg | 1.90 Å | 2023-07-19 | X-ray | DTT, MLA, NA | SARS-CoV-2 | NSP3: PLpro | 17.28% | 20.65% | 13.20% | 2,3-DIHYDROXY-1,4-DITHIOBUTANE; SODIUM ION; MALONIC ACID | 2,3-DIHYDROXY-1,4-DITHIOBUTANE; MALONIC ACID; SODIUM ION | XDS | 99.90% | 14.8 | SSRF (BL18U1) | BUSTER | 2022-06-29 | - | ZN; NA | - | - | - | N/A | - | - | Papain-like protease nsp3 | - | - | No | - | - | 76.9 | - | - | - | ||||||
| 7yh6 | 3.40 Å | 2023-07-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-07-12 | 37452031 | - | - | - | - | N/A | - | - | Spike protein S1; NIV-8 Fab heavy chain; NIV-8 Fab light chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7yh7 | 3.30 Å | 2023-07-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-07-13 | 37452031 | - | - | - | - | N/A | - | - | Spike glycoprotein; NIV-8 Fab light chain; NIV-8 Fab heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7yhw | 3.09 Å | 2023-07-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-07-14 | 37479708 | ZN | - | - | - | N/A | - | - | Spike protein S1 - RBD; Angiotensin-converting enzyme 2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7yj3 | 3.14 Å | 2023-07-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-07-19 | 37479708 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2; Spike protein S1 - RBD | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7yv8 | 2.94 Å | 2023-07-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-08-18 | 37479708 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme; Spike glycoprotein - RBD | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 7yvu | 3.20 Å | 2023-07-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-08-19 | 37479708 | ZN | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike protein S1 - RBD | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8el2 | 2.89 Å | 2023-07-19 | X-ray | NAG | SARS-CoV-2 | Spike | 23.87% | 28.15% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.90% | 11.7 | APS (23-ID-D) | PHENIX | 2022-09-22 | 37531329 | ZN | - | - | - | N/A | - | - | Fab ICO-hu23 Heavy Chain; Fab ICO-hu23 Light Chain; Spike protein S1 | - | - | No | - | 13.4 | - | - | - | |||||||
| 8elj | 3.60 Å | 2023-07-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-09-24 | 37531329 | - | - | - | - | N/A | - | - | ICO-hu23 antibody Fab heavy chain; ICO-hu23 antibody Fab light chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8gry | 3.29 Å | 2023-07-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-09-03 | 37479708 | ZN | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike protein S1 - RBD | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8h06 | 2.66 Å | 2023-07-19 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-09-28 | 37479708 | ZN | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike protein S1 - RBD | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8h0i | 2.80 Å | 2023-07-19 | Cryo-EM | - | HIV-1 | Spike | N/A | N/A | N/A | RNA (5'-R(*CP*GP*GP*UP*UP*GP*AP*UP*CP*GP*UP*UP*UP*UP*AP*AP*CP*AP*A)-3') | - | - | - | () | PHENIX | 2022-09-29 | 37419875 | ZN | - | - | - | N/A | - | - | RNA (5'-R(*CP*GP*GP*UP*UP*GP*AP*UP*CP*GP*UP*UP*UP*UP*AP*AP*CP*AP*A)-3'); APOBEC3G; Viral infectivity factor; Core binding factor beta | N/A | - | No | - | N/A | N/A | N/A | RNA (5'-R(*CP*GP*GP*UP*UP*GP*AP*UP*CP*GP*UP*UP*UP*UP*AP*AP*CP*AP*A)-3') | - | |||||||
| 8h5c | 2.90 Å | 2023-07-19 | X-ray | NAG | SARS-CoV-2 | Spike | 19.67% | 24.24% | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 97.00% | 12.1 | SSRF (BL02U1) | PHENIX | 2022-10-12 | 37479708 | ZN | - | - | - | N/A | - | - | Spike protein S1; Processed angiotensin-converting enzyme 2 | - | - | No | - | 42.6 | - | - | - | |||||||
| 8j62 | 2.50 Å | 2023-07-19 | Cryo-EM | - | HIV-1 | Spike | N/A | N/A | N/A | RNA (5'-R(*CP*GP*GP*UP*UP*GP*AP*UP*UP*GP*UP*UP*UP*UP*AP*AP*CP*AP*A)-3') | - | - | - | () | PHENIX | 2023-04-24 | 37419875 | ZN | - | - | - | N/A | - | - | RNA (5'-R(*CP*GP*GP*UP*UP*GP*AP*UP*UP*GP*UP*UP*UP*UP*AP*AP*CP*AP*A)-3'); APOBEC3G; Viral infectivity factor; Core binding factor beta | N/A | - | No | - | N/A | N/A | N/A | RNA (5'-R(*CP*GP*GP*UP*UP*GP*AP*UP*UP*GP*UP*UP*UP*UP*AP*AP*CP*AP*A)-3') | - | |||||||
| 7ur9 | 2.16 Å | 2023-07-26 | X-ray | O5F | SARS-CoV-2 | NSP5 (3CLpro) | 16.06% | 19.99% | 11.90% | (2P)-2-(isoquinolin-4-yl)-1-[4-(methylamino)-4-oxobutyl]-N-[(1S)-1-(naphthalen-2-yl)ethyl]-1H-benzimidazole-7-carboxamide | MOSFLM | 99.95% | 8.2 | ALS (8.2.1) | PHENIX | 2022-04-21 | 37542196 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 81.8 | - | - | - | ||||||||
| 7urb | 2.14 Å | 2023-07-26 | X-ray | O5O | SARS-CoV-2 | NSP5 (3CLpro) | 20.04% | 25.40% | N/A | (2P)-2-(isoquinolin-4-yl)-1-[(1s,3R)-3-(methylcarbamoyl)cyclobutyl]-N-{(1S)-1-[4-(trifluoromethyl)phenyl]butyl}-1H-benzimidazole-7-carboxamide | (2P)-2-(isoquinolin-4-yl)-1-[(1s,3R)-3-(methylcarbamoyl)cyclobutyl]-N-{(1S)-1-[4-(trifluoromethyl)phenyl]butyl}-1H-benzimidazole-7-carboxamide | iMOSFLM | 91.32% | 7.64 | ALS (8.2.1) | PHENIX | 2022-04-21 | 37542196 | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | 31.6 | - | - | - | |||||||
| 7us4 | 2.07 Å | 2023-07-26 | X-ray | O69 | SARS-CoV-2 | NSP5 (3CLpro) | 18.09% | 23.53% | N/A | (2P)-2-(isoquinolin-4-yl)-1-[(1s,3R)-3-(methylcarbamoyl)cyclobutyl]-N-[(1S)-1-(naphthalen-2-yl)ethyl]-1H-benzimidazole-7-carboxamide | MOSFLM | 99.85% | 14.43 | ALS (8.2.1) | PHENIX | 2022-04-22 | 37542196 | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | 49.6 | - | - | - | ||||||||
| 8cmb | 1.84 Å | 2023-07-26 | X-ray | DHL | SARS-CoV-2 | Spike | 19.31% | 23.12% | 12.60% | 2-AMINO-ETHANETHIOL; Spike protein S2' | 2-AMINO-ETHANETHIOL | XDS | 100.00% | 7.71 | Diamond (I04) | PHENIX | 2023-02-19 | 37471227 | - | - | - | - | N/A | - | - | Human leukocyte antigen DR beta chain allotype DR1 (DRB1*0101); Spike protein S2'; HLA class II histocompatibility antigen, DR alpha chain | - | - | No | - | 53.8 | - | - | - | |||||||
| 8cmc | 1.42 Å | 2023-07-26 | X-ray | MES | SARS-CoV-2 | Spike | 18.64% | 20.85% | 11.60% | Spike protein S2'; 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID | DIALS | 99.98% | 16.93 | Diamond (I04) | PHENIX | 2023-02-19 | 37471227 | - | - | - | - | N/A | - | - | Human leukocyte antigen DR beta chain allotype DR1 (DRB1*0101); Spike protein S2'; HLA class II histocompatibility antigen, DR alpha chain | - | - | No | - | 75.3 | - | - | - | |||||||
| 8frj | 1.57 Å | 2023-07-26 | X-ray | AW2 | SARS-CoV-2 | NSP14 | 16.98% | 20.43% | 10.50% | 5-bromo-7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine | 5-bromo-7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine | XDS | 94.30% | 10.7 | NSLS-II (17-ID-2) | PHENIX | 2023-01-07 | 37523415 | ZN | - | - | - | N/A | - | - | Transcription factor ETV6,Guanine-N7 methyltransferase nsp14 chimera | - | - | No | - | 78.6 | - | - | - | |||||||
| 8frk | 1.61 Å | 2023-07-26 | X-ray | EOH, MJ7, K | SARS-CoV-2 | NSP14 | 21.72% | 25.11% | 6.20% | ETHANOL; POTASSIUM ION; 5'-S-(4-{[(4'-chloro[1,1'-biphenyl]-3-yl)methyl]amino}butyl)-5'-thioadenosine | ETHANOL; 5'-S-(4-{[(4'-chloro[1,1'-biphenyl]-3-yl)methyl]amino}butyl)-5'-thioadenosine; POTASSIUM ION | XDS | 94.30% | 9.3 | NSLS-II (17-ID-1) | PHENIX | 2023-01-07 | 37523415 | ZN; K | - | - | - | N/A | - | - | Transcription factor ETV6,Guanine-N7 methyltransferase nsp14 chimera | - | - | No | - | 34.1 | - | - | - | |||||||
| 7wq8 | 2.20 Å | 2023-08-02 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 17.82% | 22.21% | 6.70% | Z-DEVD-FMK | - | HKL-2000 | 98.83% | 19.69 | NSRRC (BL15A1) | REFMAC | 2022-01-24 | - | - | - | - | - | N/A | - | - | 3C-like proteinase; Z-DEVD-FMK | - | - | No | - | - | 63.0 | - | - | - | ||||||
| 7wq9 | 2.05 Å | 2023-08-02 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 20.97% | 24.63% | 5.90% | Z-IETD-FMK | - | HKL-2000 | 99.50% | 20.47 | NSRRC (BL15A1) | REFMAC | 2022-01-25 | - | - | - | - | - | N/A | - | - | Z-IETD-FMK; 3C-like proteinase | - | - | No | - | - | 38.8 | - | - | - | ||||||
| 7wqb | 1.87 Å | 2023-08-02 | X-ray | ALD | SARS-CoV-2 | NSP5 (3CLpro) | 18.78% | 22.77% | 4.40% | N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | HKL-2000 | 99.60% | 29.27 | NSRRC (TPS 05A) | PHENIX | 2022-01-25 | - | MG | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 57.5 | - | - | - | ||||||
| 7wqk | 2.15 Å | 2023-08-02 | X-ray | ALD | SARS-CoV-2 | NSP5 (3CLpro) | 22.81% | 28.92% | 4.60% | N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | HKL-2000 | 98.20% | 28.582 | NSRRC (BL15A1) | REFMAC | 2022-01-25 | - | MG | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 10.2 | - | - | - | ||||||
| 7y71 | 3.12 Å | 2023-08-02 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-06-21 | 37494438 | - | - | - | - | N/A | - | - | Fab E7 heavy chain; Fab E7 light chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7y72 | 4.03 Å | 2023-08-02 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-06-21 | 37494438 | - | - | - | - | N/A | - | - | Fab E7 heavy chain; Fab E7 light chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8dpz | 3.38 Å | 2023-08-02 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-07-18 | - | - | - | - | - | N/A | - | - | DH1338 Fab Heavy Chain; DH1338 Fab Light Chain; Spike protein S1 | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 8dtk | 3.77 Å | 2023-08-02 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-07-25 | - | - | - | - | - | N/A | - | - | DH1047 Fab Light Chain; DH1047 Fab Heavy Chain; Spike protein S1 | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 8ohn | 3.40 Å | 2023-08-02 | Cryo-EM | NAG | HKU1 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2023-03-21 | 37794193 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8opm | 3.70 Å | 2023-08-02 | Cryo-EM | NAG | HKU1 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2023-04-07 | 37794193 | - | - | - | - | N/A | - | - | Spike glycoprotein,General control transcription factor GCN4 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8opn | 4.70 Å | 2023-08-02 | Cryo-EM | NAG | HKU1 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2023-04-07 | 37794193 | - | - | - | - | N/A | - | - | Spike glycoprotein,General control transcription factor GCN4 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8opo | 3.60 Å | 2023-08-02 | Cryo-EM | NAG | HKU1 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2023-04-07 | 37794193 | - | - | - | - | N/A | - | - | Spike glycoprotein,General control transcription factor GCN4 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8sph | 2.71 Å | 2023-08-02 | X-ray | NAG | SARS-CoV-2 | Spike | 21.88% | 28.29% | 15.20% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | autoPROC | 85.50% | 5.3 | APS (24-ID-C) | PHENIX | 2023-05-03 | 37578233 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2 - UNP residues 19-615; Spike protein S1 - receptor-binding domain | - | - | No | - | 12.9 | - | - | - | |||||||
| 8spi | 3.06 Å | 2023-08-02 | X-ray | NAG | SARS-CoV-2 | Spike | 18.99% | 24.35% | 12.10% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | autoPROC | 86.80% | 6.8 | APS (24-ID-E) | PHENIX | 2023-05-03 | 37578233 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme 2 - UNP residues 19-615; Spike protein S1 - receptor-binding domain | - | - | No | - | 41.6 | - | - | - | |||||||
| 7y20 | 3.80 Å | 2023-08-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-06-09 | 37376697 | - | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7y21 | 2.80 Å | 2023-08-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-06-09 | 37376697 | - | - | - | - | N/A | - | - | Processed angiotensin-converting enzyme 2; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7ybh | 3.50 Å | 2023-08-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-06-29 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7ybl | 3.60 Å | 2023-08-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-06-29 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7ybm | 3.45 Å | 2023-08-09 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-06-29 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7ymt | 6.55 Å | 2023-08-09 | Cryo-EM | NAG | MERS-CoV | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-07-29 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7ymv | 6.74 Å | 2023-08-09 | Cryo-EM | NAG | MERS-CoV | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-07-29 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7ymw | 6.05 Å | 2023-08-09 | Cryo-EM | NAG | MERS-CoV | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-07-29 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7ymx | 4.44 Å | 2023-08-09 | Cryo-EM | NAG | MERS-CoV | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-07-29 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7ymy | 4.96 Å | 2023-08-09 | Cryo-EM | NAG | MERS-CoV | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-07-29 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7ymz | 4.39 Å | 2023-08-09 | Cryo-EM | NAG | MERS-CoV | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-07-29 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7yn0 | 4.10 Å | 2023-08-09 | Cryo-EM | NAG | MERS-CoV | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-07-29 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 8b56 | 1.82 Å | 2023-08-09 | X-ray | OZI, BR | SARS-CoV-2 | NSP5 (3CLpro) | 18.49% | 23.14% | 14.20% | BROMIDE ION; (2~{S})-4-(2-chloranylethanoyl)-1-(3,4-dichlorophenyl)-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide | (2~{S})-4-(2-chloranylethanoyl)-1-(3,4-dichlorophenyl)-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide; BROMIDE ION | XDS | 64.10% | 11.4 | PETRA III, EMBL c/o DESY (P14 (MX2)) | PHENIX | 2022-09-21 | 36475694 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 53.6 | - | - | - | |||||||
| 8fiv | 2.51 Å | 2023-08-09 | X-ray | Y0I | SARS-CoV-2 | NSP5 (3CLpro) | 21.04% | 27.71% | 93.10% | (3Z)-N-([1,1'-biphenyl]-4-yl)-3-imino-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]propanamide | (3Z)-N-([1,1'-biphenyl]-4-yl)-3-imino-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]propanamide | HKL-3000 | 92.40% | 7.7 | APS (19-ID) | REFMAC | 2022-12-16 | 37482021 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 15.5 | - | - | - | |||||||
| 8fiw | 2.54 Å | 2023-08-09 | X-ray | Y1E, Y0E | SARS-CoV-2 | NSP5 (3CLpro) | 21.82% | 26.27% | 96.20% | N-([1,1'-biphenyl]-4-yl)-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]prop-2-enamide; N-([1,1'-biphenyl]-4-yl)-N-[(1S)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]prop-2-enamide | N-([1,1'-biphenyl]-4-yl)-N-[(1S)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]prop-2-enamide; N-([1,1'-biphenyl]-4-yl)-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]prop-2-enamide | HKL-3000 | 92.80% | 6.7 | APS (19-ID) | REFMAC | 2022-12-16 | 37482021 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 24.7 | - | - | - | |||||||
| 8tbe | 2.15 Å | 2023-08-09 | X-ray | ZQB | SARS-CoV-2 | NSP5 (3CLpro) | 22.91% | 29.86% | 20.00% | Pomotrelvir bound form | Pomotrelvir bound form | XDS | 98.80% | 5.1 | ALBA (XALOC) | REFMAC | 2023-06-28 | 37800975 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 7.6 | - | - | - | |||||||
| 7yrz | 1.79 Å | 2023-08-16 | X-ray | 4WI | H-CoV-229E | NSP5 (3CLpro) | 19.15% | 21.96% | 3.60% | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | XDS | 99.10% | 13.7 | SSRF (BL02U1) | PHENIX | 2022-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 65.6 | - | - | - | ||||||
| 8a95 | 2.64 Å | 2023-08-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-06-27 | 36574772 | - | - | - | - | N/A | - | - | Fab47 (Heavy chain variable domain); Fab47 (Light chain variable domain); SARS-CoV2 Trimeric Spike in 1-up conformation in complex with three Fab47 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8aiq | 2.19 Å | 2023-08-16 | X-ray | MXU | SARS-CoV-2 | NSP5 (3CLpro) | 16.82% | 22.29% | 13.30% | ~{tert}-butyl ~{N}-[1-[(2~{S})-1-[[(2~{S},3~{R})-4-azanyl-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-3-cyclopropyl-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | ~{tert}-butyl ~{N}-[1-[(2~{S})-1-[[(2~{S},3~{R})-4-azanyl-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-3-cyclopropyl-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | XDS | 98.60% | 10.9 | PETRA III, DESY (P11) | REFMAC | 2022-07-27 | - | - | - | - | - | N/A | - | - | Replicase polyprotein 1ab | - | - | No | - | - | 62.4 | - | - | - | ||||||
| 8aiu | 2.00 Å | 2023-08-16 | X-ray | M9X | SARS-CoV-2 | NSP5 (3CLpro) | 16.05% | 20.60% | N/A | tert-butyl N-[1-[(2S)-3-cyclopropyl-1-[[(2S,3R)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | tert-butyl N-[1-[(2S)-3-cyclopropyl-1-[[(2S,3R)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | XDS | 95.70% | 14.9 | PETRA III, DESY (P11) | REFMAC | 2022-07-27 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 77.3 | - | - | - | ||||||
| 8aiv | 2.60 Å | 2023-08-16 | X-ray | MFL | SARS-CoV-2 | NSP5 (3CLpro) | 18.15% | 25.17% | N/A | tert-butyl N-[1-[(2S)-3-cyclopropyl-1-[[(2S,3R)-4-(methylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | tert-butyl N-[1-[(2S)-3-cyclopropyl-1-[[(2S,3R)-4-(methylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | XDS | 98.30% | 8.0 | PETRA III, DESY (P11) | REFMAC | 2022-07-27 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 33.8 | - | - | - | ||||||
| 8aiz | 1.99 Å | 2023-08-16 | X-ray | MIJ | SARS-CoV-2 | NSP5 (3CLpro) | 17.84% | 22.39% | N/A | (2~{R},3~{S})-3-[[(2~{S})-3-cyclopropyl-2-[2-oxidanylidene-3-(2-phenylethanoylamino)pyridin-1-yl]propanoyl]amino]-~{N}-methyl-2-oxidanyl-4-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butanamide | (2~{R},3~{S})-3-[[(2~{S})-3-cyclopropyl-2-[2-oxidanylidene-3-(2-phenylethanoylamino)pyridin-1-yl]propanoyl]amino]-~{N}-methyl-2-oxidanyl-4-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butanamide | XDS | 98.90% | 13.1 | PETRA III, DESY (P11) | REFMAC | 2022-07-27 | - | - | - | - | - | N/A | - | - | Replicase polyprotein 1ab | - | - | No | - | - | 61.4 | - | - | - | ||||||
| 8aj0 | 2.52 Å | 2023-08-16 | X-ray | NA, MJ0 | SARS-CoV-2 | NSP5 (3CLpro) | 18.56% | 26.61% | N/A | (2R,3S)-3-[[(2S)-3-cyclopropyl-2-[2-oxidanylidene-3-(3-phenylpropanoylamino)pyridin-1-yl]propanoyl]amino]-N-methyl-2-oxidanyl-4-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butanamide; SODIUM ION | SODIUM ION; (2R,3S)-3-[[(2S)-3-cyclopropyl-2-[2-oxidanylidene-3-(3-phenylpropanoylamino)pyridin-1-yl]propanoyl]amino]-N-methyl-2-oxidanyl-4-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butanamide | XDS | 98.60% | 6.4 | PETRA III, DESY (P11) | REFMAC | 2022-07-27 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 22.1 | - | - | - | ||||||
| 8aja | 2.59 Å | 2023-08-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-07-27 | 37845250 | - | - | - | - | N/A | - | - | Spike glycoprotein,Fibritin | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8ajl | 2.77 Å | 2023-08-16 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-07-28 | 37845250 | - | - | - | - | N/A | - | - | Spike glycoprotein,Fibritin | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8hr2 | 1.94 Å | 2023-08-16 | X-ray | - | SARS-CoV-2 | Spike | 19.08% | 22.21% | N/A | - | 100.00% | 24.7 | SSRF (BL19U1) | PHENIX | 2022-12-14 | 37419228 | - | - | - | - | N/A | - | - | NB1C6; Spike protein S1 - Receptor-binding domain (RBD); NB1B5 | - | - | No | - | 63.0 | - | - | - | |||||||||
| 8i30 | 2.00 Å | 2023-08-16 | X-ray | OF9 | SARS-CoV-2 | NSP5 (3CLpro) | 18.66% | 22.62% | 4.20% | (2~{R})-1-[4,4-bis(fluoranyl)cyclohexyl]carbonyl-4,4-bis(fluoranyl)-~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(pyridin-2-ylmethylamino)butan-2-yl]pyrrolidine-2-carboxamide | (2~{R})-1-[4,4-bis(fluoranyl)cyclohexyl]carbonyl-4,4-bis(fluoranyl)-~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(pyridin-2-ylmethylamino)butan-2-yl]pyrrolidine-2-carboxamide | XDS | 97.50% | 13.7 | SSRF (BL18U1) | PHENIX | 2023-01-16 | 37517202 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 58.9 | - | - | - | |||||||
| 8iv4 | 3.59 Å | 2023-08-16 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2023-03-26 | 37470320 | - | - | - | - | N/A | - | - | heavy chain of 3E2; light chain of 8H12; heavy chain of 8H12; Spike protein S1; light chain of 3E2 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 8iv5 | 3.77 Å | 2023-08-16 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2023-03-26 | 37470320 | - | - | - | - | N/A | - | - | heavy chain of 8H12; light chain of 8H12; Spike protein S1; heavy chain of 1C4; light chain of 1C4 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 8iv8 | 3.92 Å | 2023-08-16 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2023-03-26 | 37470320 | - | - | - | - | N/A | - | - | Spike protein S1; heavy chain of 3E2; light chain of 1C4; light chain of 3E2; heavy chain of 1C4 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 8iva | 3.95 Å | 2023-08-16 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2023-03-26 | 37470320 | - | - | - | - | N/A | - | - | heavy chain of XMA01; Spike protein S1; light chain of 3E2; heavy chain of 3E2; light chain of XMA01 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 8jop | 2.70 Å | 2023-08-16 | X-ray | UWO | SARS-CoV-2 | NSP5 (3CLpro) | 19.47% | 24.82% | 8.80% | methyl (6~{R})-5-ethanoyl-7-oxidanylidene-6-[4-(trifluoromethyl)phenyl]-8,9,10,11-tetrahydro-6~{H}-benzo[b][1,4]benzodiazepine-2-carboxylate | methyl (6~{R})-5-ethanoyl-7-oxidanylidene-6-[4-(trifluoromethyl)phenyl]-8,9,10,11-tetrahydro-6~{H}-benzo[b][1,4]benzodiazepine-2-carboxylate | XDS | 99.70% | 11.6 | SSRF (BL19U1) | PHENIX | 2023-06-08 | 37437852 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 36.9 | - | - | - | |||||||
| 8sk5 | 2.01 Å | 2023-08-16 | X-ray | - | SARS-CoV-2 | Spike | 18.32% | 21.09% | 7.10% | - | autoPROC | 70.90% | 18.3 | APS (17-ID) | PHENIX | 2023-04-18 | 37608223 | - | - | - | - | N/A | - | - | Spike protein S1 - receptor binding domain; anti-SARS-CoV-2 receptor binding domain VHH | - | - | No | - | 73.4 | - | - | - | ||||||||
| 8gf2 | 2.85 Å | 2023-08-23 | X-ray | NAG | SARS-CoV-2 | Spike | 20.90% | 25.18% | 14.50% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.40% | 4.7 | SSRL (BL12-1) | PHENIX | 2023-03-07 | 37582161 | - | - | - | - | N/A | - | - | eCR3022.20 Fab light chain; CC12.3 Fab heavy chain; CC12.3 Fab light chain; eCR3022.20 Fab heavy chain; Spike protein S1 | - | - | No | - | 33.7 | - | - | - | |||||||
| 8gw4 | 2.90 Å | 2023-08-23 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 22.10% | 24.36% | N/A | peptide 8-1 | - | CrysalisPro | 99.18% | 19.7 | ROTATING ANODE () | PHENIX | 2022-09-16 | - | - | - | - | - | N/A | - | - | Replicase polyprotein 1ab - UNP residues 3264-3565; peptide 8-1 | - | - | No | - | - | 41.5 | - | - | - | ||||||
| 8gws | 2.90 Å | 2023-08-23 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 22.21% | 25.25% | N/A | VAL-LYS-LEU-GLN-ALA-ILE-PHE-ARG | - | CrysalisPro | 99.01% | 23.0 | ROTATING ANODE () | PHENIX | 2022-09-17 | - | - | - | - | - | N/A | - | - | VAL-LYS-LEU-GLN-ALA-ILE-PHE-ARG; Replicase polyprotein 1ab - UNP residues 3264-3565 | - | - | No | - | - | 33.0 | - | - | - | ||||||
| 8jpq | 2.70 Å | 2023-08-23 | X-ray | ACY, ALA, V3E, ILE | SARS-CoV-2 | NSP5 (3CLpro) | 19.92% | 24.97% | 7.12% | ALANINE; methyl (2~{S})-2-azanyl-3-(2-hydroxyphenyl)propanoate; ISOLEUCINE | ACETIC ACID; ALANINE; methyl (2~{S})-2-azanyl-3-(2-hydroxyphenyl)propanoate; ISOLEUCINE | 99.68% | 22.61 | ROTATING ANODE () | PHENIX | 2023-06-12 | - | - | - | - | - | N/A | - | - | ORF1a polyprotein | - | - | No | - | - | 35.6 | - | - | - | |||||||
| 8cyu | 1.80 Å | 2023-08-30 | X-ray | P5X | SARS-CoV-2 | NSP5 (3CLpro) | 21.21% | 24.53% | 13.20% | N-[(4-chlorothiophen-2-yl)methyl]-N-[4-(dimethylamino)phenyl]-2-(isoquinolin-4-yl)acetamide | N-[(4-chlorothiophen-2-yl)methyl]-N-[4-(dimethylamino)phenyl]-2-(isoquinolin-4-yl)acetamide | xia2 | 99.90% | 8.5 | CLSI (08B1-1) | REFMAC | 2022-05-24 | 37555275 | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | 39.8 | - | - | - | |||||||
| 8cyz | 1.90 Å | 2023-08-30 | X-ray | P6I | SARS-CoV-2 | NSP5 (3CLpro) | 24.47% | 27.76% | 18.50% | N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)-N-[4-(methylsulfanyl)phenyl]acetamide | N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)-N-[4-(methylsulfanyl)phenyl]acetamide | xia2 | 99.80% | 7.1 | CLSI (08B1-1) | REFMAC | 2022-05-24 | 37555275 | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | 15.3 | - | - | - | |||||||
| 8cz4 | 2.10 Å | 2023-08-30 | X-ray | P6R | SARS-CoV-2 | NSP5 (3CLpro) | 21.60% | 24.38% | 10.00% | N-(4-tert-butylphenyl)-N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)acetamide | N-(4-tert-butylphenyl)-N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)acetamide | CrysalisPro | 96.10% | 3.2 | ROTATING ANODE () | REFMAC | 2022-05-24 | 37555275 | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | 41.4 | - | - | - | |||||||
| 8cz7 | 2.00 Å | 2023-08-30 | X-ray | P7L | SARS-CoV-2 | NSP5 (3CLpro) | 24.19% | 27.11% | N/A | N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)-N-(4-methoxyphenyl)acetamide | N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)-N-(4-methoxyphenyl)acetamide | CrysalisPro | 94.60% | 6.62 | ROTATING ANODE () | REFMAC | 2022-05-24 | 37555275 | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | 18.8 | - | - | - | |||||||
| 8e7t | 2.50 Å | 2023-08-30 | X-ray | - | MERS-CoV | NSP5 (3CLpro) | 18.59% | 24.41% | 19.09% | SODIUM ION | SODIUM ION | CrysalisPro | 99.86% | 10.61 | ROTATING ANODE () | PHENIX | 2022-08-24 | - | - | - | - | - | N/A | - | - | 3C-Like Protease | - | - | No | - | - | 40.9 | - | - | - | ||||||
| 8fy6 | 2.00 Å | 2023-08-30 | X-ray | OW1 | SARS-CoV-2 | NSP5 (3CLpro) | 18.74% | 22.17% | 23.20% | Nirmatrelvir (reacted form) | Nirmatrelvir (reacted form) | xia2 | 93.80% | 5.3 | APS (23-ID-B) | PHENIX | 2023-01-25 | 37595260 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 63.5 | - | - | - | |||||||
| 8fy7 | 1.94 Å | 2023-08-30 | X-ray | YFK | SARS-CoV-2 | NSP5 (3CLpro) | 17.06% | 21.35% | 7.40% | 4-methoxy-N-[(2S)-4-methyl-1-oxo-1-({(2S)-1-[(3S)-2-oxopyrrolidin-3-yl]but-3-en-2-yl}amino)pentan-2-yl]-1H-indole-2-carboxamide | 4-methoxy-N-[(2S)-4-methyl-1-oxo-1-({(2S)-1-[(3S)-2-oxopyrrolidin-3-yl]but-3-en-2-yl}amino)pentan-2-yl]-1H-indole-2-carboxamide | xia2 | 100.00% | 12.5 | APS (23-ID-D) | PHENIX | 2023-01-25 | 37595260 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 71.1 | - | - | - | |||||||
| 8gnh | 3.74 Å | 2023-08-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-08-23 | - | - | - | - | - | N/A | - | - | Heavy chain of BD-218; Light chain of BD-218; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 8gqt | 2.09 Å | 2023-08-30 | X-ray | QUE | SARS-CoV-2 | NSP5 (3CLpro) | 20.54% | 25.32% | 15.50% | 3,5,7,3',4'-PENTAHYDROXYFLAVONE | 3,5,7,3',4'-PENTAHYDROXYFLAVONE | XDS | 98.60% | 5.28 | SSRF (BL19U1) | PHENIX | 2022-08-30 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 32.3 | - | - | - | ||||||
| 8h7l | 2.44 Å | 2023-08-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-10-20 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; BA7535 fab light chain; BA7535 fab heavt chain | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 8h7z | 3.07 Å | 2023-08-30 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-10-21 | - | - | - | - | - | N/A | - | - | BA7535 fab; BA7535 fab; Spike glycoprotein - RBD | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 8hfx | 2.98 Å | 2023-08-30 | Cryo-EM | NAG | HIV-1/SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose; beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-11-13 | 37676003 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme; Spike glycoprotein,Envelope glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8hfy | 3.21 Å | 2023-08-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-11-13 | 37676003 | ZN | - | - | - | N/A | - | - | Spike protein S1; Angiotensin-converting enzyme | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8hfz | 2.71 Å | 2023-08-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-11-13 | 37676003 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8hg0 | 3.51 Å | 2023-08-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-11-13 | 37676003 | ZN | - | - | - | N/A | - | - | Spike protein S1; Angiotensin-converting enzyme | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8htv | 2.04 Å | 2023-08-30 | X-ray | UZF | SARS-CoV-2 | NSP5 (3CLpro) | 18.37% | 23.63% | 99.50% | 1-(5,6-dihydrobenzo[b][1]benzazepin-11-yl)-2-sulfanyl-ethanone | 1-(5,6-dihydrobenzo[b][1]benzazepin-11-yl)-2-sulfanyl-ethanone | HKL-3000 | 99.50% | 5.6 | SSRF (BL18U1) | PHENIX | 2022-12-21 | 37594952 | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | 48.6 | - | - | - | |||||||
| 8ify | 2.55 Å | 2023-08-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2023-02-20 | 37676003 | ZN | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8ifz | 2.85 Å | 2023-08-30 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2023-02-20 | 37676003 | ZN | - | - | - | N/A | - | - | Angiotensin-converting enzyme; Spike protein S1 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8sty | 1.90 Å | 2023-08-30 | X-ray | WGE | SARS-CoV-2 | NSP5 (3CLpro) | 21.46% | 25.04% | 7.80% | benzyl (3S)-3-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamoyl)-2-azaspiro[4.4]nonane-2-carboxylate | benzyl (3S)-3-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamoyl)-2-azaspiro[4.4]nonane-2-carboxylate | PROTEUM PLUS | 98.93% | 12.9 | ROTATING ANODE () | PHENIX | 2023-05-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 34.8 | - | - | - | ||||||
| 8stz | 1.85 Å | 2023-08-30 | X-ray | WGI | SARS-CoV-2 | NSP5 (3CLpro) | 20.19% | 22.90% | N/A | benzyl (3S)-3-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamoyl)-2-azaspiro[4.5]decane-2-carboxylate | benzyl (3S)-3-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamoyl)-2-azaspiro[4.5]decane-2-carboxylate | PROTEUM PLUS | 99.80% | 13.9 | ROTATING ANODE () | PHENIX | 2023-05-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 55.9 | - | - | - | ||||||
| 8sxr | 2.11 Å | 2023-08-30 | X-ray | WZK | SARS-CoV-2 | NSP5 (3CLpro) | 19.92% | 25.19% | N/A | N-[(4-chlorothiophen-2-yl)methyl]-N-[4-(dimethylamino)phenyl]-2-(5-hydroxyisoquinolin-4-yl)acetamide | XDS | 67.50% | 11.241 | CLSI (08B1-1) | REFMAC | 2023-05-23 | 37555275 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 33.7 | - | - | - | ||||||||
| 7u92 | 1.80 Å | 2023-09-06 | X-ray | M0C | SARS-CoV-2 | NSP5 (3CLpro) | 17.19% | 20.74% | N/A | (1R,2S,5S)-N-{(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | XDS | 99.50% | 9.6 | SSRL (BL12-2) | REFMAC | 2022-03-09 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 76.2 | - | - | - | ||||||
| 7y0o | 4.40 Å | 2023-09-06 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2022-06-05 | - | - | - | - | - | N/A | - | - | Spike protein S1 - RBD; P14 nanobody; light chain of R15; heavy chain of R15 | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||||
| 7ybk | 3.90 Å | 2023-09-06 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-06-29 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7ylb | 2.41 Å | 2023-09-06 | X-ray | - | SARS-CoV-2 | Nucleocapsid | 26.71% | 32.59% | 18.60% | - | HKL-3000 | 99.80% | 7.6 | SSRF (BL19U1) | REFMAC | 2022-07-26 | - | - | - | - | - | N/A | - | - | Nucleoprotein - CTD; NC2 | - | - | No | - | - | 4.6 | - | - | - | |||||||
| 8gsb | 3.99 Å | 2023-09-06 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-09-05 | - | - | - | - | - | N/A | - | - | Light chain of VacBB-665; Spike protein S1; heavy chain of VacBB-665 | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 7xy3 | 4.60 Å | 2023-09-13 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-05-31 | - | - | - | - | - | N/A | - | - | VHH14; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||||
| 7xy4 | 3.30 Å | 2023-09-13 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-05-31 | - | - | - | - | - | N/A | - | - | VHH21; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||||
| 7y0n | 3.80 Å | 2023-09-13 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-06-05 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; P14 Nanobody; Light chain of R15-F7; Heavy chain of R15-F7 | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 8azc | 0.93 Å | 2023-09-13 | X-ray | MPO | SARS-CoV-2 | NSP3: Macro | 14.26% | 15.31% | N/A | 3[N-MORPHOLINO]PROPANE SULFONIC ACID | 3[N-MORPHOLINO]PROPANE SULFONIC ACID | XDS | 98.57% | 15.51 | PETRA III, EMBL c/o DESY (P13 (MX1)) | PHENIX | 2022-09-05 | - | - | - | - | - | N/A | - | - | Papain-like protease nsp3 | - | - | No | - | - | 98.0 | - | - | - | ||||||
| 8azd | 2.00 Å | 2023-09-13 | X-ray | APR | SARS-CoV-2 | NSP3: Macro | 21.29% | 25.04% | N/A | ADENOSINE-5-DIPHOSPHORIBOSE | ADENOSINE-5-DIPHOSPHORIBOSE | XDS | 99.39% | 9.2 | PETRA III, EMBL c/o DESY (P13 (MX1)) | PHENIX | 2022-09-05 | - | - | - | - | - | N/A | - | - | Papain-like protease nsp3 | - | - | No | - | - | 34.8 | - | - | - | ||||||
| 8f0i | 3.70 Å | 2023-09-13 | X-ray | NAG | SARS-CoV-2 | Spike | 22.58% | 27.50% | 95.50% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 99.80% | 1.9 | SSRL (BL12-2) | PHENIX | 2022-11-03 | 37841584 | - | - | - | - | N/A | - | - | Spike protein S1 - Receptor binding domain, UNP residues 333-530; COVA309-22 heavy chain; COVA309-22 light chain | - | - | No | - | 16.6 | - | - | - | |||||||
| 8gtv | 1.80 Å | 2023-09-13 | X-ray | KAE | SARS-CoV-2 | NSP5 (3CLpro) | 22.39% | 24.69% | 9.10% | 4-[(2~{S})-4-(3,4-dichlorophenyl)-2-(morpholin-4-ylmethyl)piperazin-1-yl]carbonyl-1~{H}-quinolin-2-one | 4-[(2~{S})-4-(3,4-dichlorophenyl)-2-(morpholin-4-ylmethyl)piperazin-1-yl]carbonyl-1~{H}-quinolin-2-one | HKL-2000 | 99.80% | 3.8 | SSRF (BL19U1) | PHENIX | 2022-09-08 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 38.4 | - | - | - | ||||||
| 8gtw | 1.85 Å | 2023-09-13 | X-ray | K9U | SARS-CoV-2 | NSP5 (3CLpro) | 19.81% | 22.74% | 16.70% | (2S)-4-(3,4-dichlorophenyl)-1-[(2-oxidanylidene-1H-quinolin-4-yl)carbonyl]-N-[3,3,3-tris(fluoranyl)propyl]piperazine-2-carboxamide | (2S)-4-(3,4-dichlorophenyl)-1-[(2-oxidanylidene-1H-quinolin-4-yl)carbonyl]-N-[3,3,3-tris(fluoranyl)propyl]piperazine-2-carboxamide | HKL-2000 | 99.90% | 4.2 | SSRF (BL19U1) | PHENIX | 2022-09-08 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 57.7 | - | - | - | ||||||
| 8u2x | 2.25 Å | 2023-09-13 | X-ray | TRS | SARS-CoV-2 | NSP15 | 18.70% | 21.33% | 18.20% | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL | XDS | 100.00% | 14.5 | NSLS-II (19-ID) | PHENIX | 2023-09-06 | - | - | - | - | - | N/A | - | - | Uridylate-specific endoribonuclease nsp15 | - | - | No | - | - | 71.2 | - | - | - | ||||||
| 7yr4 | 4.12 Å | 2023-09-20 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2022-08-08 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme 2 | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||||
| 8edf | 3.40 Å | 2023-09-20 | X-ray | NAG | SARS-CoV-2 | Spike | 27.59% | 29.94% | 28.70% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 96.60% | 5.8 | SSRL (BL12-1) | PHENIX | 2022-09-04 | 37722054 | - | - | - | - | N/A | - | - | Spike protein S1; SKD Fab Light chain; SKD Fab heavy chain | - | - | No | - | 7.4 | - | - | - | |||||||
| 8gvd | 2.00 Å | 2023-09-20 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 18.60% | 24.81% | N/A | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE | - | CrysalisPro | 99.70% | 12.36 | ROTATING ANODE () | REFMAC | 2022-09-14 | - | - | - | - | - | N/A | - | - | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE; 3C-like proteinase nsp5 | - | - | No | - | - | 37.0 | - | - | - | ||||||
| 8gvy | 2.50 Å | 2023-09-20 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 21.46% | 27.10% | N/A | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE | - | CrysalisPro | 99.51% | 9.83 | ROTATING ANODE () | PHENIX | 2022-09-16 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5; 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE | - | - | No | - | - | 18.9 | - | - | - | ||||||
| 8gwj | 2.90 Å | 2023-09-20 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 21.14% | 27.06% | N/A | VAL-LYS-LEU-GLN-ALA-VAL-PHE-ARG | - | CrysalisPro | 98.20% | 33.87 | ROTATING ANODE () | REFMAC | 2022-09-17 | - | - | - | - | - | N/A | - | - | VAL-LYS-LEU-GLN-ALA-VAL-PHE-ARG; Replicase polyprotein 1ab - UNP residues 3264-3565 | - | - | No | - | - | 19.2 | - | - | - | ||||||
| 8dd6 | 2.30 Å | 2023-09-27 | X-ray | K36 | SARS-CoV-2 | NSP5 (3CLpro) | 19.73% | 25.04% | 17.50% | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | DIALS | 99.96% | 5.8 | CHESS (F1) | PHENIX | 2022-06-17 | 37699927 | - | - | - | - | N/A | - | - | ORF1a polyprotein | - | - | No | - | 34.8 | - | - | - | |||||||
| 8gxg | 1.69 Å | 2023-09-27 | X-ray | 06Q | SARS-CoV-2 | NSP5 (3CLpro) | 16.65% | 20.61% | 4.60% | N-[(2S)-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2S,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | N-[(2S)-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2S,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | XDS | 98.80% | 21.27 | SSRF (BL19U1) | PHENIX | 2022-09-20 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 77.2 | - | - | - | ||||||
| 8gxh | 1.59 Å | 2023-09-27 | X-ray | 0AX | SARS-CoV-2 | NSP5 (3CLpro) | 17.02% | 19.37% | 3.30% | N-[(2S)-3-cyclohexyl-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | N-[(2S)-3-cyclohexyl-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | XDS | 99.40% | 25.1 | SSRF (BL19U1) | PHENIX | 2022-09-20 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 85.8 | - | - | - | ||||||
| 8gxi | 1.69 Å | 2023-09-27 | X-ray | 0BO | SARS-CoV-2 | NSP5 (3CLpro) | 16.57% | 20.32% | 4.70% | N-[(2S)-3-cyclohexyl-1-[[(2S,3R)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-1-benzofuran-2-carboxamide | N-[(2S)-3-cyclohexyl-1-[[(2S,3R)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-1-benzofuran-2-carboxamide | XDS | 98.80% | 18.05 | SSRF (BL19U1) | PHENIX | 2022-09-20 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 79.4 | - | - | - | ||||||
| 8gzb | 2.70 Å | 2023-09-27 | X-ray | KM6 | SARS-CoV-2 | NSP5 (3CLpro) | 20.78% | 25.64% | 16.60% | 2-(4-chlorophenyl)-1,3,4-oxadiazole | 2-(4-chlorophenyl)-1,3,4-oxadiazole | XDS | 99.90% | 8.4 | SSRF (BL17B1) | PHENIX | 2022-09-26 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 29.6 | - | - | - | ||||||
| 8jsl | 2.95 Å | 2023-09-27 | Cryo-EM | - | EBOV | Spike | N/A | N/A | N/A | The leader sequence of EBOV | - | - | - | () | 2023-06-20 | 37699521 | ZN | - | - | - | N/A | - | - | Polymerase cofactor VP35; The leader sequence of EBOV; RNA-directed RNA polymerase L | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8jsn | 3.40 Å | 2023-09-27 | Cryo-EM | - | EBOV | Spike | N/A | N/A | N/A | The leader sequence of EBOV genome | - | - | - | () | 2023-06-20 | 37699521 | ZN | - | - | - | N/A | - | - | RNA-directed RNA polymerase L; Polymerase cofactor VP35; The leader sequence of EBOV genome | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8p99 | 3.40 Å | 2023-09-27 | Cryo-EM | NAG | SARS-CoV-2 | Spike | 42.27% | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2023-06-05 | 37471688 | - | - | - | - | N/A | - | - | Spike protein S1,Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8p9y | 4.30 Å | 2023-09-27 | Cryo-EM | NAG, XIO, NA | SARS-CoV-2 | Spike | 53.38% | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; [(2~{S})-2-[[4-(2-azanylethanoylamino)-7-[[(2~{S})-3-[2-(4-nitrophenyl)sulfanyl-1~{H}-indol-3-yl]-1-oxidanylidene-1-sodiooxy-propan-2-yl]amino]-4-[3-[[(2~{S})-3-[2-(4-nitrophenyl)sulfanyl-1~{H}-indol-3-yl]-1-oxidanylidene-1-sodiooxy-propan-2-yl]amino]-3-oxidanylidene-propyl]-7-oxidanylidene-heptanoyl]amino]-3-[2-(4-nitrophenyl)sulfanyl-1~{H}-indol-3-yl]propanoyl]oxysodium; SODIUM ION | 2-acetamido-2-deoxy-beta-D-glucopyranose; [(2~{S})-2-[[4-(2-azanylethanoylamino)-7-[[(2~{S})-3-[2-(4-nitrophenyl)sulfanyl-1~{H}-indol-3-yl]-1-oxidanylidene-1-sodiooxy-propan-2-yl]amino]-4-[3-[[(2~{S})-3-[2-(4-nitrophenyl)sulfanyl-1~{H}-indol-3-yl]-1-oxidanylidene-1-sodiooxy-propan-2-yl]amino]-3-oxidanylidene-propyl]-7-oxidanylidene-heptanoyl]amino]-3-[2-(4-nitrophenyl)sulfanyl-1~{H}-indol-3-yl]propanoyl]oxysodium; SODIUM ION | - | - | () | 2023-06-06 | 37471688 | - | - | - | - | N/A | - | - | Spike protein S1,Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8sg6 | 2.49 Å | 2023-09-27 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 19.27% | 24.00% | 14.40% | - | DIALS | 100.00% | 8.6 | CHESS (F1) | PHENIX | 2023-04-11 | 37699927 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 44.7 | - | - | - | ||||||||
| 7ukl | 3.09 Å | 2023-10-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-04-01 | 37776849 | - | - | - | - | N/A | - | - | 12-16 Fab Heavy Chain; Spike glycoprotein; 12-16 Fab Light Chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7ukm | 3.03 Å | 2023-10-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-04-01 | - | - | - | - | - | N/A | - | - | 12-19 Fab Heavy Chain; 12-19 Fab Light Chain; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 8crf | 1.15 Å | 2023-10-04 | X-ray | NT9 | SARS-CoV-2 | NSP1 | 19.17% | 24.46% | N/A | ~{N}-methyl-1-(4-thiophen-2-ylphenyl)methanamine | ~{N}-methyl-1-(4-thiophen-2-ylphenyl)methanamine | xia2 | 100.00% | 63.7 | Diamond, Diamond, ESRF (I23, I23, MASSIF-1) | PHENIX | 2023-03-08 | 37446375 | - | - | - | - | N/A | - | - | Host translation inhibitor nsp1 | - | - | No | - | 40.5 | - | - | - | |||||||
| 8crk | 1.10 Å | 2023-10-04 | X-ray | OG3 | SARS-CoV-2 | NSP1 | 16.75% | 19.18% | N/A | (1~{R})-1-(4-chlorophenyl)ethanamine | (1~{R})-1-(4-chlorophenyl)ethanamine | xia2 | 96.90% | 34.3 | Diamond, Diamond, ESRF (I23, I23, MASSIF-1) | PHENIX | 2023-03-08 | 37446375 | - | - | - | - | N/A | - | - | Host translation inhibitor nsp1 | - | - | No | - | 86.8 | - | - | - | |||||||
| 8crm | 1.42 Å | 2023-10-04 | X-ray | OF6 | SARS-CoV-2 | NSP1 | 17.85% | 20.28% | N/A | 1-[2-(3-chlorophenyl)-1,3-thiazol-4-yl]-~{N}-methyl-methanamine | 1-[2-(3-chlorophenyl)-1,3-thiazol-4-yl]-~{N}-methyl-methanamine | xia2 | 99.60% | 23.2 | Diamond, Diamond, ESRF (I23, I23, MASSIF-1) | PHENIX | 2023-03-08 | 37446375 | - | - | - | - | N/A | - | - | Host translation inhibitor nsp1 | - | - | No | - | 79.8 | - | - | - | |||||||
| 8fr7 | 3.39 Å | 2023-10-04 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2023-01-06 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||||
| 8fxb | 3.10 Å | 2023-10-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2023-01-24 | 37648855 | - | - | - | - | N/A | - | - | S309 Heavy chain; Processed angiotensin-converting enzyme 2 - ectodomain; Spike protein S1 - Receptor binding domain; S309 Light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8fxc | 3.20 Å | 2023-10-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2023-01-24 | 37648855 | ZN | - | - | - | N/A | - | - | S309 Light chain; Spike protein S1 - Receptor binding domain; Processed angiotensin-converting enzyme 2 - ectodomain; S309 Heavy chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8i3s | 3.90 Å | 2023-10-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2023-01-17 | 37704615 | - | - | - | - | N/A | - | - | Light chain of Fab 7B3; Heavy chain od Fab 7B3; Spike glycoprotein - RBD region | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8i3u | 3.60 Å | 2023-10-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2023-01-18 | 37704615 | - | - | - | - | N/A | - | - | Spike glycoprotein - RBD region; Heavy chain of Fab 14B1; Light chain of Fab 14B1 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8s9g | 3.00 Å | 2023-10-04 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2023-03-28 | 37648855 | - | - | - | - | N/A | - | - | Spike glycoprotein - RBD; S309 Fab Heavy chain; Processed angiotensin-converting enzyme 2 - ectodomain; S309 Fab Light chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8sak | 3.00 Å | 2023-10-04 | Cryo-EM | NAG | MERS-CoV | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2023-04-01 | 37756379 | - | - | - | - | N/A | - | - | JC57-11 Fab heavy chain; JC57-11 Fab light chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 7uug | 2.00 Å | 2023-10-11 | X-ray | M0C | SARS-CoV-2 | NSP5 (3CLpro) | 18.24% | 24.04% | N/A | (1R,2S,5S)-N-{(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | XDS | 99.90% | 13.2 | SSRL (BL12-2) | REFMAC | 2022-04-28 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 44.5 | - | - | - | ||||||
| 7uup | 2.00 Å | 2023-10-11 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | 19.14% | 23.72% | N/A | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | XDS | 99.20% | 7.8 | SSRL (BL12-2) | REFMAC | 2022-04-28 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 47.6 | - | - | - | ||||||
| 8bwu | 2.36 Å | 2023-10-11 | X-ray | 6NR | SARS-CoV-2 | NSP14 | 23.00% | 26.39% | 37.83% | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid | XDS | 99.46% | 7.63 | BESSY (14.1) | PHENIX | 2022-12-07 | - | ZN | - | - | - | N/A | - | - | Transcription factor ETV6,Proofreading exoribonuclease nsp14 | - | - | No | - | - | 23.9 | - | - | - | ||||||
| 8eoy | 2.28 Å | 2023-10-11 | X-ray | WOH | SARS-CoV-2 | NSP5 (3CLpro) | 23.63% | 29.92% | 18.70% | benzyl {(2S)-1-[2-(3-amino-3-oxopropyl)-2-(chloroacetyl)hydrazinyl]-4-methyl-1-oxopentan-2-yl}carbamate (non-preferred name) | benzyl {(2S)-1-[2-(3-amino-3-oxopropyl)-2-(chloroacetyl)hydrazinyl]-4-methyl-1-oxopentan-2-yl}carbamate (non-preferred name) | 97.90% | 5.7 | ROTATING ANODE () | PHENIX | 2022-10-04 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 7.4 | - | - | - | |||||||
| 8eqf | 3.16 Å | 2023-10-11 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-10-07 | - | - | - | - | - | N/A | - | - | Fab heavy chain; Fab light chain; Spike protein S1 | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 8ezv | 1.80 Å | 2023-10-11 | X-ray | X6O | SARS-CoV-2 | NSP5 (3CLpro) | 17.88% | 22.00% | 14.50% | (1R,2S,5S)-N-{(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopiperidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | XDS | 99.60% | 8.0 | SSRL (BL12-2) | REFMAC | 2022-11-01 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 65.0 | - | - | - | |||||||
| 8ezz | 1.85 Å | 2023-10-11 | X-ray | X70 | SARS-CoV-2 | NSP5 (3CLpro) | 19.04% | 25.40% | 9.00% | (1R,2S,5S)-N-{(2S,3R)-4-(3,3-difluoroazetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopiperidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(2S,3R)-4-(3,3-difluoroazetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopiperidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | DIALS | 98.80% | 7.2 | SSRL (BL12-2) | REFMAC | 2022-11-01 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 31.6 | - | - | - | ||||||
| 8f02 | 2.00 Å | 2023-10-11 | X-ray | X6T | SARS-CoV-2 | NSP5 (3CLpro) | 18.97% | 25.48% | N/A | (1R,2S,5S)-N-{(2S,3R)-4-(3,3-dimethylazetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopiperidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(2S,3R)-4-(3,3-dimethylazetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopiperidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | XDS | 97.90% | 5.5 | SSRL (BL12-2) | REFMAC | 2022-11-01 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 31.1 | - | - | - | ||||||
| 8f2c | 1.95 Å | 2023-10-11 | X-ray | X9Z | SARS-CoV-2 | NSP5 (3CLpro) | 19.55% | 26.68% | N/A | (1R,2S,5S)-N-[(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-(2-oxopyrrolidin-1-yl)butan-2-yl]-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-[(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-(2-oxopyrrolidin-1-yl)butan-2-yl]-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | XDS | 99.10% | 7.1 | SSRL (BL12-2) | REFMAC | 2022-11-07 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 21.8 | - | - | - | ||||||
| 8f2d | 1.95 Å | 2023-10-11 | X-ray | XA8 | SARS-CoV-2 | NSP5 (3CLpro) | 19.62% | 25.21% | 15.00% | (1R,2S,5S)-N-[(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-(2-oxopiperidin-1-yl)butan-2-yl]-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-[(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-(2-oxopiperidin-1-yl)butan-2-yl]-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | DIALS | 97.60% | 5.5 | SSRL (BL12-2) | REFMAC | 2022-11-07 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 33.2 | - | - | - | ||||||
| 8h3g | 1.46 Å | 2023-10-11 | X-ray | 7YY | SARS-CoV-2 | NSP5 (3CLpro) | 14.13% | 16.22% | 5.20% | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | 96.00% | 18.94 | SSRF (BL02U1) | PHENIX | 2022-10-08 | 37696289 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 96.7 | - | - | - | ||||||||
| 8h3k | 1.80 Å | 2023-10-11 | X-ray | VIB, 7YY | SARS-CoV-2 | NSP5 (3CLpro) | 16.81% | 20.13% | 7.90% | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM; 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM; 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | 97.40% | 15.29 | SSRF (BL02U1) | PHENIX | 2022-10-08 | 37696289 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 80.8 | - | - | - | ||||||||
| 8h3l | 2.30 Å | 2023-10-11 | X-ray | 7YY | SARS-CoV-2 | NSP5 (3CLpro) | 20.64% | 23.07% | 13.20% | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | 99.40% | 7.55 | SSRF (BL02U1) | PHENIX | 2022-10-08 | 37696289 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 54.4 | - | - | - | ||||||||
| 8h4y | 2.25 Å | 2023-10-11 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | 20.57% | 24.76% | 8.60% | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | autoPROC | 99.60% | 18.95 | Diamond (I04) | PHENIX | 2022-10-11 | 37696289 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 37.6 | - | - | - | |||||||
| 8h51 | 2.18 Å | 2023-10-11 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | 18.76% | 23.43% | 10.80% | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | autoPROC | 99.80% | 18.0 | Diamond (I04) | PHENIX | 2022-10-11 | 37696289 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 50.7 | - | - | - | |||||||
| 8h57 | 1.55 Å | 2023-10-11 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | 17.72% | 20.38% | 6.90% | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | autoPROC | 99.10% | 22.61 | Diamond (I04) | PHENIX | 2022-10-12 | 37696289 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 79.1 | - | - | - | |||||||
| 8h5f | 1.79 Å | 2023-10-11 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | 18.40% | 22.73% | 8.00% | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | autoPROC | 99.90% | 21.05 | Diamond (I04) | PHENIX | 2022-10-13 | 37696289 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 57.8 | - | - | - | |||||||
| 8h5p | 1.67 Å | 2023-10-11 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | 18.03% | 20.75% | 5.90% | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | autoPROC | 99.70% | 25.0 | Diamond (I04) | PHENIX | 2022-10-13 | 37696289 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 76.2 | - | - | - | |||||||
| 8h6i | 1.90 Å | 2023-10-11 | X-ray | ODN | SARS-CoV-2 | NSP5 (3CLpro) | 19.78% | 22.73% | 7.50% | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one | autoPROC | 98.80% | 12.15 | SSRF (BL02U1) | PHENIX | 2022-10-17 | 37696289 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 57.8 | - | - | - | |||||||
| 8h6n | 1.65 Å | 2023-10-11 | X-ray | 4WI, LQZ | SARS-CoV-2 | NSP5 (3CLpro) | 19.30% | 21.60% | 5.80% | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide; 2-(diethylamino)-N-(2,6-dimethylphenyl)ethanamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide; 2-(diethylamino)-N-(2,6-dimethylphenyl)ethanamide | autoPROC | 99.80% | 25.42 | Diamond (I04) | PHENIX | 2022-10-18 | 37696289 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 68.8 | - | - | - | |||||||
| 8h7k | 1.45 Å | 2023-10-11 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 16.81% | 18.38% | 3.40% | - | autoPROC | 99.70% | 24.02 | SSRF (BL02U1) | PHENIX | 2022-10-20 | 37696289 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5; nsp4/5 peptidyl substrate | - | - | No | - | 90.7 | - | - | - | ||||||||
| 8h7w | 1.60 Å | 2023-10-11 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | 16.81% | 19.65% | 10.40% | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | autoPROC | 99.80% | 18.73 | Diamond (I04) | PHENIX | 2022-10-21 | 37696289 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 84.0 | - | - | - | |||||||
| 8h82 | 1.93 Å | 2023-10-11 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | 17.00% | 21.56% | 10.60% | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | autoPROC | 99.80% | 17.65 | Diamond (I04) | PHENIX | 2022-10-21 | 37696289 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 69.3 | - | - | - | |||||||
| 8hbk | 1.80 Å | 2023-10-11 | X-ray | 7YY | SARS-CoV-2 | NSP5 (3CLpro) | 19.11% | 23.22% | 4.70% | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | autoPROC | 97.80% | 20.48 | SSRF (BL02U1) | PHENIX | 2022-10-29 | 37696289 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 52.8 | - | - | - | |||||||
| 8f4s | 2.15 Å | 2023-10-18 | X-ray | NA, FMT, XDU | SARS-CoV-2 | NSP10/NSP16 | 16.56% | 19.09% | 17.80% | FORMIC ACID; 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol; SODIUM ION | SODIUM ION; FORMIC ACID; 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol | HKL-3000 | 99.90% | 24.4 | APS (21-ID-D) | REFMAC | 2022-11-11 | 37728236 | ZN; NA | - | - | - | N/A | - | - | Non-structural protein 10; 2'-O-methyltransferase | - | - | No | - | 87.3 | - | - | - | |||||||
| 8f4y | 2.13 Å | 2023-10-18 | X-ray | XE0, NA, XDU, FMT | SARS-CoV-2 | NSP10/NSP16 | 15.73% | 17.98% | 10.40% | 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol; FORMIC ACID; 4-[2-(2,4-dichlorophenyl)ethyl]-6-(trifluoromethyl)pyrimidin-2-ol; SODIUM ION | 4-[2-(2,4-dichlorophenyl)ethyl]-6-(trifluoromethyl)pyrimidin-2-ol; SODIUM ION; 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol; FORMIC ACID | HKL-3000 | 99.90% | 17.5 | APS (21-ID-D) | REFMAC | 2022-11-11 | 37728236 | ZN; NA | - | - | - | N/A | - | - | 2'-O-methyltransferase; Non-structural protein 10 | - | - | No | - | 92.2 | - | - | - | |||||||
| 8h5t | 2.00 Å | 2023-10-18 | X-ray | NAG | SARS-CoV-2 | Spike | 17.11% | 20.47% | 11.00% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-3000 | 100.00% | 46.136 | SSRF (BL19U1) | PHENIX | 2022-10-13 | - | - | - | - | - | N/A | - | - | Spike protein S1; Nanobody Nb-015 | - | - | No | - | - | 78.4 | - | - | - | ||||||
| 8h5u | 2.40 Å | 2023-10-18 | X-ray | NAG | SARS-CoV-2 | Spike | 22.52% | 24.51% | 17.30% | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | HKL-2000 | 97.90% | 25.692 | SSRF (BL19U1) | PHENIX | 2022-10-13 | - | - | - | - | - | N/A | - | - | Spike protein S1; Nanobody Nb-021 | - | - | No | - | - | 39.9 | - | - | - | ||||||
| 8ifp | 1.78 Å | 2023-10-18 | X-ray | OZ6 | SARS-CoV-2 | NSP5 (3CLpro) | 26.51% | 29.18% | 8.00% | (1R,2S,5S)-3-[(2S)-2-(tert-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-6,6-dimethyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-3-[(2S)-2-(tert-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-6,6-dimethyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | XDS | 97.50% | 15.8 | SSRF (BL19U1) | PHENIX | 2023-02-19 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 9.4 | - | - | - | ||||||
| 8ifq | 1.96 Å | 2023-10-18 | X-ray | I1Z | SARS-CoV-2 | NSP5 (3CLpro) | 17.92% | 21.42% | 9.60% | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide | XDS | 95.40% | 14.5 | SSRF (BL19U1) | PHENIX | 2023-02-19 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 70.4 | - | - | - | ||||||
| 8ifr | 1.66 Å | 2023-10-18 | X-ray | P0O | SARS-CoV-2 | NSP5 (3CLpro) | 18.12% | 21.57% | 13.10% | (1R,2S,5S)-3-[(2S)-2-(tert-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-6,6-dimethyl-N-[(2S)-5-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]hex-3-en-2-yl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-3-[(2S)-2-(tert-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-6,6-dimethyl-N-[(2S)-5-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]hex-3-en-2-yl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | XDS | 99.10% | 13.2 | SSRF (BL02U1) | PHENIX | 2023-02-19 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 69.2 | - | - | - | ||||||
| 8ifs | 2.46 Å | 2023-10-18 | X-ray | OZL | SARS-CoV-2 | NSP5 (3CLpro) | 26.63% | 29.55% | 10.10% | (8~{S})-7-[(2~{S})-2-(~{tert}-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-~{N}-[(1~{S})-1-cyano-2-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]ethyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide | (8~{S})-7-[(2~{S})-2-(~{tert}-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-~{N}-[(1~{S})-1-cyano-2-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]ethyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide | XDS | 97.80% | 9.4 | SSRF (BL19U1) | PHENIX | 2023-02-19 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 8.4 | - | - | - | ||||||
| 8ift | 1.80 Å | 2023-10-18 | X-ray | OZB | SARS-CoV-2 | NSP5 (3CLpro) | 18.07% | 20.54% | 98.20% | (8S)-N-[(1S)-1-cyano-2-[(3S)-2-oxidanylidenepyrrolidin-3-yl]ethyl]-7-[(2S)-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide | (8S)-N-[(1S)-1-cyano-2-[(3S)-2-oxidanylidenepyrrolidin-3-yl]ethyl]-7-[(2S)-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide | HKL-2000 | 100.00% | 3.3 | SSRF (BL19U1) | PHENIX | 2023-02-19 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 77.8 | - | - | - | ||||||
| 8igx | 1.90 Å | 2023-10-18 | X-ray | PQL | SARS-CoV-2 | NSP5 (3CLpro) | 17.60% | 21.25% | 99.10% | (8~{S})-~{N}-[(1~{S})-1-cyano-2-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]ethyl]-7-[(2~{S})-3,3-dimethyl-2-[2,2,2-tris(fluoranyl)ethanoylamino]butanoyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide | (8~{S})-~{N}-[(1~{S})-1-cyano-2-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]ethyl]-7-[(2~{S})-3,3-dimethyl-2-[2,2,2-tris(fluoranyl)ethanoylamino]butanoyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide | HKL-2000 | 99.70% | 2.5 | SSRF (BL18U1) | PHENIX | 2023-02-21 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 72.0 | - | - | - | ||||||
| 8igy | 1.96 Å | 2023-10-18 | X-ray | 4WI | SARS-CoV-2 | NSP5 (3CLpro) | 18.74% | 20.56% | 11.50% | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide | XDS | 92.20% | 15.1 | SSRF (BL17U1) | PHENIX | 2023-02-21 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 77.7 | - | - | - | ||||||
| 8ppk | 2.98 Å | 2023-10-18 | Cryo-EM | UNX | Bat-Hp-CoV | NSP1 | N/A | N/A | N/A | UNKNOWN ATOM OR ION | UNKNOWN ATOM OR ION | - | - | () | 2023-07-07 | 37802027 | MG; ZN | - | - | - | N/A | - | - | 40S ribosomal protein S10; 40S ribosomal protein S9; 40S ribosomal protein S8; 40S ribosomal protein S7; Eukaryotic translation initiation factor 1; 40S ribosomal protein S11; 40S ribosomal protein S6; 40S ribosomal protein S5; 40S ribosomal protein S4, X isoform; 40S ribosomal protein S3; 40S ribosomal protein S2; 40S ribosomal protein S3a; Nsp1; 60S ribosomal protein L41; Receptor of activated protein C kinase 1; Ubiquitin-40S ribosomal protein S27a; Small ribosomal subunit protein eS30; 40S ribosomal protein S29; 40S ribosomal protein S28; 40S ribosomal protein S27; 40S ribosomal protein SA; 40S ribosomal protein S26; 40S ribosomal protein S25; 40S ribosomal protein S24; 40S ribosomal protein S23; 40S ribosomal protein S15a; 40S ribosomal protein S21; 40S ribosomal protein S20; Small ribosomal subunit protein eS19; Small ribosomal subunit protein uS13; 40S ribosomal protein S17; 18S rRNA; 40S ribosomal protein S16; 40S ribosomal protein S15; 40S ribosomal protein S14; 40S ribosomal protein S13; 40S ribosomal protein S12 | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8ufl | 2.51 Å | 2023-10-18 | X-ray | - | SARS-CoV | NSP3(SUD-M) | 21.35% | 26.72% | 11.20% | - | HKL-3000 | 100.00% | 15.1 | APS (21-ID-F) | REFMAC | 2023-10-04 | - | - | - | - | - | N/A | - | - | Papain-like protease nsp3 - SARS-Unique Domain (SUD) | - | - | No | - | - | 21.4 | - | - | - | |||||||
| 8ufm | 1.65 Å | 2023-10-18 | X-ray | FMT | SARS-CoV-2 | NSP3(SUD-M) | 18.48% | 20.79% | 10.00% | FORMIC ACID | FORMIC ACID | HKL-3000 | 99.90% | 30.1 | APS (21-ID-D) | REFMAC | 2023-10-04 | - | - | - | - | - | N/A | - | - | Papain-like protease nsp3 - SARS-Unique Domain (SUD) | - | - | No | - | - | 75.9 | - | - | - | ||||||
| 8gw1 | 3.31 Å | 2023-10-25 | Cryo-EM | MN | SARS-CoV-2 | NSP7/NSP8/NSP9/NSP12/NSP13 | N/A | N/A | N/A | MANGANESE (II) ION | MANGANESE (II) ION | - | - | () | PHENIX | 2022-09-16 | 36335936 | MN; ZN | - | - | - | N/A | - | - | Non-structural protein 8 - UNP residues 3943-4140; Replicase polyprotein 1ab - UNP residues 4393-5324; Non-structural protein 7 - UNP residues 3860-3942; RNA (25-MER); Template; Non-structural protein 9; Helicase - UNP residues 5325-5925 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8h6f | 3.30 Å | 2023-10-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose; 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | phenix.real_space_refine; PHENIX | 2022-10-17 | - | - | - | - | - | N/A | - | - | Repebody (A6); Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 8hgl | 2.90 Å | 2023-10-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-11-15 | 37452031 | - | - | - | - | N/A | - | - | NIV-11 Fab light chain; NIV-11 Fab heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||
| 8hgm | 3.40 Å | 2023-10-25 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | PHENIX | 2022-11-15 | 37452031 | - | - | - | - | N/A | - | - | NIV-11 Fab light chain; NIV-11 Fab heavy chain; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8hi9 | 2.28 Å | 2023-10-25 | X-ray | LKR | SARS-CoV-2 | NSP5 (3CLpro) | 24.57% | 25.53% | 6.70% | 3,7-bis(oxidanyl)-2-[3,4,5-tris(oxidanyl)phenyl]chromen-4-one | XDS | 99.60% | 14.8 | SSRF (BL19U1) | PHENIX | 2022-11-19 | 37259339 | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | 30.5 | - | - | - | ||||||||
| 8suz | - | 2023-10-25 | NMR | - | SARS-CoV-2 | Envelope | N/A | N/A | N/A | - | - | - | () | 2023-05-14 | 37831764 | - | - | - | - | N/A | - | - | Envelope small membrane protein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 8t20 | 3.36 Å | 2023-10-25 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2023-06-05 | 37842796 | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 8t21 | 3.60 Å | 2023-10-25 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2023-06-05 | 37842796 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 8t22 | 3.83 Å | 2023-10-25 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2023-06-05 | 37842796 | - | - | - | - | N/A | - | - | Spike glycoprotein; Angiotensin-converting enzyme | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 8t23 | 3.87 Å | 2023-10-25 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2023-06-05 | 37842796 | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme; Spike glycoprotein - RBD | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8t25 | 3.62 Å | 2023-10-25 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2023-06-05 | 37842796 | - | - | - | - | N/A | - | - | Spike glycoprotein - RBD; Angiotensin-converting enzyme | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 8taz | 3.75 Å | 2023-10-25 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2023-06-28 | 37842796 | - | - | - | - | N/A | - | - | Angiotensin-converting enzyme; Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 8bec | 1.70 Å | 2023-11-01 | X-ray | - | SARS-CoV-2 | Spike | 18.54% | 20.68% | 7.89% | - | XDS | 99.96% | 17.68 | SOLEIL (PROXIMA 1) | PHENIX | 2022-10-21 | - | - | - | - | - | N/A | - | - | pT1375 single-chain Fv; Spike protein S1 | - | - | No | - | - | 76.8 | - | - | - | |||||||
| 8eyg | 3.73 Å | 2023-11-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-10-27 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; Nanobody | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 8eyh | 3.75 Å | 2023-11-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-10-27 | - | - | - | - | - | N/A | - | - | Nanobody; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 8gy6 | - | 2023-11-01 | Cryo-EM | GO3 | SARS-CoV-2 | NSP7/NSP8/NSP12 | N/A | N/A | N/A | Gossypol | Gossypol | - | - | () | 2022-09-21 | - | - | - | - | - | N/A | - | - | RNA-directed RNA polymerase; Non-structural protein 7; Non-structural protein 8 | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 8h91 | 3.07 Å | 2023-11-01 | X-ray | - | SARS-CoV-2 | Spike | 21.17% | 28.42% | 5.40% | - | XDS | 98.90% | 7.6 | SSRF (BL17U1) | PHENIX | 2022-10-24 | - | - | - | - | - | N/A | - | - | nanobody; Spike protein S1 | - | - | No | - | - | 12.2 | - | - | - | |||||||
| 8pq2 | 3.85 Å | 2023-11-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2023-07-10 | 37852477 | - | - | - | - | N/A | - | - | Spike protein S2'; P4J15 Fragment Antigen-Binding Light Chain; P4J15 Fragment Antigen-Binding Heavy Chain | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 8psd | 2.90 Å | 2023-11-01 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2023-07-13 | 37852477 | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | N/A | N/A | N/A | - | |||||||||
| 7gav | 1.77 Å | 2023-11-08 | X-ray | KFU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.60% | 23.63% | 24.00% | (3S)-5-chloro-N-(isoquinolin-4-yl)-2,3-dihydro-1-benzofuran-3-carboxamide | (3S)-5-chloro-N-(isoquinolin-4-yl)-2,3-dihydro-1-benzofuran-3-carboxamide | MOSFLM | 99.90% | 7.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 48.6 | - | - | - | |||||
| 7gaw | 1.81 Å | 2023-11-08 | X-ray | KG9 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.41% | 25.20% | 32.30% | (4S)-6-chloro-2-[(1-cyanocyclopropyl)methanesulfonyl]-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-2-[(1-cyanocyclopropyl)methanesulfonyl]-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 99.90% | 5.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 33.3 | - | - | - | |||||
| 7gax | 1.71 Å | 2023-11-08 | X-ray | KJI | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.82% | 21.95% | 8.20% | N-(4-methylpyridin-3-yl)-N~2~-(quinolin-4-yl)glycinamide | N-(4-methylpyridin-3-yl)-N~2~-(quinolin-4-yl)glycinamide | XDS | 99.60% | 9.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 65.6 | - | - | - | |||||
| 7gay | 1.30 Å | 2023-11-08 | X-ray | KJO | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 15.78% | 19.67% | 7.20% | N-phenyl-2-(pyridin-3-yl)prop-2-enamide | N-phenyl-2-(pyridin-3-yl)prop-2-enamide | XDS | 89.70% | 7.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 83.9 | - | - | - | |||||
| 7gaz | 1.75 Å | 2023-11-08 | X-ray | KL6 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.25% | 21.45% | 7.40% | 1-{2-[(methanesulfonyl)amino]ethyl}-1,2,3,4-tetrahydroquinoline-7-sulfonamide | 1-{2-[(methanesulfonyl)amino]ethyl}-1,2,3,4-tetrahydroquinoline-7-sulfonamide | XDS | 85.60% | 10.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 70.2 | - | - | - | |||||
| 7gb0 | 1.42 Å | 2023-11-08 | X-ray | KLR | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 15.68% | 19.94% | 5.30% | (2S)-N-tert-butyl-2-[4-(2-cyanoethyl)anilino]-2-(pyridin-3-yl)acetamide | (2S)-N-tert-butyl-2-[4-(2-cyanoethyl)anilino]-2-(pyridin-3-yl)acetamide | XDS | 98.50% | 10.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 82.1 | - | - | - | |||||
| 7gb1 | 1.29 Å | 2023-11-08 | X-ray | KMF | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 15.88% | 19.68% | 6.60% | N-[2-(4-acetylpiperazin-1-yl)ethyl]naphthalene-1-carboxamide | N-[2-(4-acetylpiperazin-1-yl)ethyl]naphthalene-1-carboxamide | XDS | 88.50% | 8.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 83.9 | - | - | - | |||||
| 7gb2 | 1.50 Å | 2023-11-08 | X-ray | KMX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.04% | 20.33% | 5.90% | 1-{4-[(2-benzyl-1,3-thiazol-5-yl)methyl]piperazin-1-yl}ethan-1-one | 1-{4-[(2-benzyl-1,3-thiazol-5-yl)methyl]piperazin-1-yl}ethan-1-one | XDS | 98.00% | 11.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 79.4 | - | - | - | |||||
| 7gb3 | 1.38 Å | 2023-11-08 | X-ray | KNU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.57% | 20.64% | 7.20% | N-[(1S)-1-(3-chloro-5-fluorophenyl)ethyl]acetamide | N-[(1S)-1-(3-chloro-5-fluorophenyl)ethyl]acetamide | XDS | 96.40% | 9.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 77.0 | - | - | - | |||||
| 7gb4 | 1.86 Å | 2023-11-08 | X-ray | KO9 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.58% | 22.99% | 8.50% | N-(5-cyanopyridin-3-yl)-2-(pyridin-3-yl)acetamide | N-(5-cyanopyridin-3-yl)-2-(pyridin-3-yl)acetamide | XDS | 98.10% | 8.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 55.2 | - | - | - | |||||
| 7gb5 | 1.26 Å | 2023-11-08 | X-ray | KOI | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.31% | 19.54% | 4.60% | 2-(3-chlorophenyl)-N-(pyridin-3-yl)acetamide | 2-(3-chlorophenyl)-N-(pyridin-3-yl)acetamide | XDS | 84.20% | 13.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 84.7 | - | - | - | |||||
| 7gb6 | 1.84 Å | 2023-11-08 | X-ray | KP0 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.62% | 21.69% | 19.80% | N-(3-chlorophenyl)-N-(2-cyclohexylethyl)-N'-(pyridin-3-yl)urea | N-(3-chlorophenyl)-N-(2-cyclohexylethyl)-N'-(pyridin-3-yl)urea | XDS | 99.80% | 3.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 68.2 | - | - | - | |||||
| 7gb7 | 1.55 Å | 2023-11-08 | X-ray | KQ3 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.88% | 20.53% | 7.30% | N-(3-chlorophenyl)-N-[2-(morpholin-4-yl)ethyl]-N'-(pyridin-3-yl)urea | N-(3-chlorophenyl)-N-[2-(morpholin-4-yl)ethyl]-N'-(pyridin-3-yl)urea | XDS | 99.40% | 10.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 77.9 | - | - | - | |||||
| 7gb8 | 1.96 Å | 2023-11-08 | X-ray | KQL | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.28% | 23.27% | 14.00% | N-(4-methylpyridin-3-yl)-2-[3-(trifluoromethyl)phenyl]acetamide | N-(4-methylpyridin-3-yl)-2-[3-(trifluoromethyl)phenyl]acetamide | XDS | 98.10% | 6.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 52.4 | - | - | - | |||||
| 7gb9 | 1.40 Å | 2023-11-08 | X-ray | KQX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.54% | 21.03% | 7.80% | 2-(4-methylphenyl)-N-(4-methylpyridin-3-yl)acetamide | 2-(4-methylphenyl)-N-(4-methylpyridin-3-yl)acetamide | XDS | 97.80% | 7.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 73.8 | - | - | - | |||||
| 7gba | 1.70 Å | 2023-11-08 | X-ray | KS0 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.66% | 25.49% | 9.40% | 1-{4-[(4-fluorophenyl)methyl]piperazin-1-yl}propan-1-one | 1-{4-[(4-fluorophenyl)methyl]piperazin-1-yl}propan-1-one | XDS | 98.00% | 7.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 31.0 | - | - | - | |||||
| 7gbb | 1.74 Å | 2023-11-08 | X-ray | KSI | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.57% | 23.45% | 9.00% | 1-[(3S)-4-[(3-chlorophenyl)methyl]-3-(2-methylpropyl)piperazin-1-yl]ethan-1-one | 1-[(3S)-4-[(3-chlorophenyl)methyl]-3-(2-methylpropyl)piperazin-1-yl]ethan-1-one | XDS | 98.30% | 9.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 50.5 | - | - | - | |||||
| 7gbc | 1.66 Å | 2023-11-08 | X-ray | KSX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.73% | 22.46% | 15.90% | 2-(5-cyanopyridin-3-yl)-N-(4-methylpyridin-3-yl)acetamide | 2-(5-cyanopyridin-3-yl)-N-(4-methylpyridin-3-yl)acetamide | XDS | 99.70% | 4.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 60.7 | - | - | - | |||||
| 7gbd | 1.78 Å | 2023-11-08 | X-ray | KT9 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.49% | 22.24% | 13.20% | N-(3-methyl-5-{[(2S)-4-oxoazetidin-2-yl]oxy}phenyl)-2-(pyrimidin-5-yl)acetamide | N-(3-methyl-5-{[(2S)-4-oxoazetidin-2-yl]oxy}phenyl)-2-(pyrimidin-5-yl)acetamide | XDS | 97.60% | 7.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 62.8 | - | - | - | |||||
| 7gbe | 1.22 Å | 2023-11-08 | X-ray | KU6 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.99% | 22.05% | 15.10% | (4R)-6-chloro-N-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 75.80% | 2.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 64.6 | - | - | - | |||||
| 7gbf | 1.77 Å | 2023-11-08 | X-ray | KUU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.47% | 21.29% | 9.10% | (2R)-2-(3-chlorophenyl)-3-methyl-N-(4-methylpyridin-3-yl)butanamide | (2R)-2-(3-chlorophenyl)-3-methyl-N-(4-methylpyridin-3-yl)butanamide | XDS | 91.70% | 7.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 71.7 | - | - | - | |||||
| 7gbg | 1.46 Å | 2023-11-08 | X-ray | KVF | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.65% | 21.25% | 7.40% | (2S)-2-(3-chlorophenyl)-N-(5-methylpyridazin-4-yl)butanamide | (2S)-2-(3-chlorophenyl)-N-(5-methylpyridazin-4-yl)butanamide | XDS | 98.60% | 8.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 72.0 | - | - | - | |||||
| 7gbh | 1.58 Å | 2023-11-08 | X-ray | KVO | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.74% | 20.93% | 7.50% | 2-(3-chlorophenyl)-N-(2,4-dimethylpyridin-3-yl)acetamide | 2-(3-chlorophenyl)-N-(2,4-dimethylpyridin-3-yl)acetamide | XDS | 99.30% | 9.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 74.6 | - | - | - | |||||
| 7gbi | 1.29 Å | 2023-11-08 | X-ray | KVX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.14% | 19.17% | 5.60% | (3S)-5-chloro-N-[4-(hydroxymethyl)pyridin-3-yl]-2,3-dihydro-1-benzofuran-3-carboxamide | (3S)-5-chloro-N-[4-(hydroxymethyl)pyridin-3-yl]-2,3-dihydro-1-benzofuran-3-carboxamide | XDS | 86.50% | 11.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 86.9 | - | - | - | |||||
| 7gbj | 1.55 Å | 2023-11-08 | X-ray | KW9 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.45% | 22.88% | 9.70% | 7-fluoro-N-[2-(2-methoxyphenoxy)ethyl]-2-oxo-1,2-dihydroquinoline-4-carboxamide | 7-fluoro-N-[2-(2-methoxyphenoxy)ethyl]-2-oxo-1,2-dihydroquinoline-4-carboxamide | XDS | 98.50% | 6.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 56.3 | - | - | - | |||||
| 7gbk | 1.50 Å | 2023-11-08 | X-ray | KXF | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.67% | 21.58% | 11.10% | 2-(3-hydroxyphenyl)-N-(4-methylpyridin-3-yl)acetamide | 2-(3-hydroxyphenyl)-N-(4-methylpyridin-3-yl)acetamide | XDS | 99.10% | 4.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 69.1 | - | - | - | |||||
| 7gbl | 2.16 Å | 2023-11-08 | X-ray | KWR | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.88% | 24.51% | 12.30% | N-(3-{[(2S)-4-oxoazetidin-2-yl]oxy}phenyl)-2-(pyridin-3-yl)acetamide | N-(3-{[(2S)-4-oxoazetidin-2-yl]oxy}phenyl)-2-(pyridin-3-yl)acetamide | XDS | 99.70% | 6.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 39.9 | - | - | - | |||||
| 7gbm | 1.80 Å | 2023-11-08 | X-ray | KX9 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.89% | 21.47% | 12.00% | (2R)-2-(3-chlorophenyl)-N-[(4M)-4-(1H-pyrazol-1-yl)pyridin-3-yl]propanamide | (2R)-2-(3-chlorophenyl)-N-[(4M)-4-(1H-pyrazol-1-yl)pyridin-3-yl]propanamide | XDS | 99.70% | 6.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 70.1 | - | - | - | |||||
| 7gbn | 1.94 Å | 2023-11-08 | X-ray | KXR | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.43% | 23.50% | 7.00% | N-(3-fluoro-5-{[(2S)-4-oxoazetidin-2-yl]oxy}phenyl)-2-(pyrimidin-5-yl)acetamide | N-(3-fluoro-5-{[(2S)-4-oxoazetidin-2-yl]oxy}phenyl)-2-(pyrimidin-5-yl)acetamide | XDS | 80.60% | 9.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 49.8 | - | - | - | |||||
| 7gbo | 1.62 Å | 2023-11-08 | X-ray | KY0 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.51% | 21.70% | 5.80% | 1-(3-chlorophenyl)-N-(4-methylpyridin-3-yl)cyclopropane-1-carboxamide | 1-(3-chlorophenyl)-N-(4-methylpyridin-3-yl)cyclopropane-1-carboxamide | XDS | 99.70% | 11.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 67.9 | - | - | - | |||||
| 7gbp | 1.70 Å | 2023-11-08 | X-ray | KYC | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.73% | 22.34% | 6.20% | (2S)-2-(3-chlorophenyl)-2-(dimethylamino)-N-(4-methylpyridin-3-yl)acetamide | (2S)-2-(3-chlorophenyl)-2-(dimethylamino)-N-(4-methylpyridin-3-yl)acetamide | XDS | 99.70% | 11.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 61.8 | - | - | - | |||||
| 7gbq | 1.72 Å | 2023-11-08 | X-ray | KYU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.22% | 22.63% | 18.70% | 2-(3-chlorophenyl)-2,2-difluoro-N-(4-methylpyridin-3-yl)acetamide | 2-(3-chlorophenyl)-2,2-difluoro-N-(4-methylpyridin-3-yl)acetamide | XDS | 95.50% | 8.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 58.8 | - | - | - | |||||
| 7gbr | 1.49 Å | 2023-11-08 | X-ray | KZC | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.40% | 25.90% | 12.90% | N-(2-anilinoethyl)-2-oxo-1,2-dihydroquinoline-4-carboxamide | N-(2-anilinoethyl)-2-oxo-1,2-dihydroquinoline-4-carboxamide | XDS | 99.00% | 3.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 27.3 | - | - | - | |||||
| 7gbs | 1.54 Å | 2023-11-08 | X-ray | KZX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.57% | 21.08% | 10.50% | 2-(3-fluorophenyl)-N-(4-methylpyridin-3-yl)acetamide | 2-(3-fluorophenyl)-N-(4-methylpyridin-3-yl)acetamide | XDS | 98.30% | 8.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 73.4 | - | - | - | |||||
| 7gbt | 1.25 Å | 2023-11-08 | X-ray | L1F | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.20% | 19.50% | 5.10% | N-[2-(2-methoxyphenoxy)ethyl]-N-methyl-2-oxo-1,2-dihydroquinoline-4-carboxamide | N-[2-(2-methoxyphenoxy)ethyl]-N-methyl-2-oxo-1,2-dihydroquinoline-4-carboxamide | XDS | 83.00% | 12.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 84.9 | - | - | - | |||||
| 7gbu | 1.56 Å | 2023-11-08 | X-ray | L2I | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.68% | 23.03% | 11.10% | (2S)-4-(methylamino)-2-phenyl-N-(pyridin-3-yl)butanamide | (2S)-4-(methylamino)-2-phenyl-N-(pyridin-3-yl)butanamide | XDS | 99.50% | 7.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 54.7 | - | - | - | |||||
| 7gbv | 1.67 Å | 2023-11-08 | X-ray | L3I | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.32% | 22.50% | 7.40% | 2-(3-chlorophenyl)-2-methyl-N-(4-methylpyridin-3-yl)propanamide | 2-(3-chlorophenyl)-2-methyl-N-(4-methylpyridin-3-yl)propanamide | XDS | 99.60% | 8.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 60.1 | - | - | - | |||||
| 7gbw | 1.56 Å | 2023-11-08 | X-ray | L6R | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.15% | 24.02% | 11.70% | (2R)-2-(3-chlorophenyl)-N-(4-methylpyridin-3-yl)pentanamide | (2R)-2-(3-chlorophenyl)-N-(4-methylpyridin-3-yl)pentanamide | XDS | 99.10% | 6.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 44.6 | - | - | - | |||||
| 7gbx | 1.91 Å | 2023-11-08 | X-ray | L6D | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.58% | 22.84% | 11.90% | N-(1H-benzimidazol-1-yl)-2-(3-chlorophenyl)acetamide | N-(1H-benzimidazol-1-yl)-2-(3-chlorophenyl)acetamide | XDS | 99.70% | 7.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 56.6 | - | - | - | |||||
| 7gby | 1.61 Å | 2023-11-08 | X-ray | L5I | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.29% | 22.22% | 10.00% | (2R)-3-cyclopropyl-2-methyl-N-(4-methylpyridin-3-yl)propanamide | (2R)-3-cyclopropyl-2-methyl-N-(4-methylpyridin-3-yl)propanamide | XDS | 96.90% | 8.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 62.9 | - | - | - | |||||
| 7gbz | 1.53 Å | 2023-11-08 | X-ray | L4U | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.88% | 22.72% | 13.40% | (3S)-3,4-dimethyl-N-(4-methylpyridin-3-yl)pentanamide | (3S)-3,4-dimethyl-N-(4-methylpyridin-3-yl)pentanamide | XDS | 99.40% | 4.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 57.9 | - | - | - | |||||
| 7gc0 | 1.58 Å | 2023-11-08 | X-ray | L4N | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.78% | 21.52% | 10.90% | (5R)-N-(4-methylpyridin-3-yl)spiro[2.4]heptane-5-carboxamide | (5R)-N-(4-methylpyridin-3-yl)spiro[2.4]heptane-5-carboxamide | XDS | 99.50% | 7.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 69.6 | - | - | - | |||||
| 7gc1 | 1.59 Å | 2023-11-08 | X-ray | L7F | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.59% | 23.91% | 16.60% | 2-[(1S,5R)-bicyclo[3.1.0]hexan-1-yl]-N-(4-methylpyridin-3-yl)acetamide | 2-[(1S,5R)-bicyclo[3.1.0]hexan-1-yl]-N-(4-methylpyridin-3-yl)acetamide | XDS | 99.10% | 4.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 45.7 | - | - | - | |||||
| 7gc2 | 1.73 Å | 2023-11-08 | X-ray | L7Q | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.82% | 23.06% | 10.40% | 3-methyl-N-(4-methylpyridin-3-yl)-3-phenylbutanamide | 3-methyl-N-(4-methylpyridin-3-yl)-3-phenylbutanamide | XDS | 99.20% | 8.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 54.5 | - | - | - | |||||
| 7gc3 | 1.56 Å | 2023-11-08 | X-ray | L7V | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.64% | 21.43% | 6.70% | 1-[(2S)-2-(5-cyclopropyl-1,2,4-oxadiazol-3-yl)pyrrolidin-1-yl]-2-(pyridin-3-yl)ethan-1-one | 1-[(2S)-2-(5-cyclopropyl-1,2,4-oxadiazol-3-yl)pyrrolidin-1-yl]-2-(pyridin-3-yl)ethan-1-one | XDS | 99.30% | 10.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 70.4 | - | - | - | |||||
| 7gc4 | 1.74 Å | 2023-11-08 | X-ray | L83 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.85% | 21.89% | 8.10% | N-(2-amino-4-methylpyridin-3-yl)-2-(3-chlorophenyl)acetamide | N-(2-amino-4-methylpyridin-3-yl)-2-(3-chlorophenyl)acetamide | XDS | 88.60% | 8.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 66.2 | - | - | - | |||||
| 7gc5 | 1.57 Å | 2023-11-08 | X-ray | L8F | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.12% | 22.73% | 9.30% | N-(2-{7-[(4-acetylpiperazin-1-yl)methyl]-1H-indol-3-yl}ethyl)acetamide | N-(2-{7-[(4-acetylpiperazin-1-yl)methyl]-1H-indol-3-yl}ethyl)acetamide | XDS | 99.40% | 7.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 57.8 | - | - | - | |||||
| 7gc6 | 1.69 Å | 2023-11-08 | X-ray | L8O | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.65% | 21.63% | 9.20% | 1-[(4R)-4-(3-methylphenyl)-3,4-dihydroisoquinolin-2(1H)-yl]ethan-1-one | 1-[(4R)-4-(3-methylphenyl)-3,4-dihydroisoquinolin-2(1H)-yl]ethan-1-one | XDS | 99.10% | 9.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 68.6 | - | - | - | |||||
| 7gc7 | 1.65 Å | 2023-11-08 | X-ray | L93 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.76% | 21.15% | 8.70% | 6-fluoro-N-[(2R)-2-(2-methoxyphenoxy)propyl]-2-oxo-1,2-dihydroquinoline-4-carboxamide | 6-fluoro-N-[(2R)-2-(2-methoxyphenoxy)propyl]-2-oxo-1,2-dihydroquinoline-4-carboxamide | XDS | 98.90% | 9.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 72.8 | - | - | - | |||||
| 7gc8 | 1.80 Å | 2023-11-08 | X-ray | L9F | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.45% | 22.84% | 8.80% | (3P,5R)-3-(3-chlorophenyl)-5-(pyridin-3-yl)imidazolidine-2,4-dione | (3P,5R)-3-(3-chlorophenyl)-5-(pyridin-3-yl)imidazolidine-2,4-dione | XDS | 99.20% | 8.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 56.6 | - | - | - | |||||
| 7gc9 | 1.77 Å | 2023-11-08 | X-ray | L9O | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.37% | 22.95% | 17.50% | (1S)-N-(4-methylpyridin-3-yl)spiro[3.3]heptane-1-carboxamide | (1S)-N-(4-methylpyridin-3-yl)spiro[3.3]heptane-1-carboxamide | XDS | 99.80% | 3.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 55.5 | - | - | - | |||||
| 7gca | 1.77 Å | 2023-11-08 | X-ray | LB0 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.85% | 21.52% | 11.90% | (1r,3r)-3-cyclopropyl-N-(4-methylpyridin-3-yl)cyclobutane-1-carboxamide | (1r,3r)-3-cyclopropyl-N-(4-methylpyridin-3-yl)cyclobutane-1-carboxamide | XDS | 99.70% | 5.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 69.6 | - | - | - | |||||
| 7gcb | 1.61 Å | 2023-11-08 | X-ray | LBC | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.90% | 22.24% | 5.70% | 2-(3-iodophenyl)-N-(4-methylpyridin-3-yl)acetamide | 2-(3-iodophenyl)-N-(4-methylpyridin-3-yl)acetamide | XDS | 99.00% | 12.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 62.8 | - | - | - | |||||
| 7gcc | 1.61 Å | 2023-11-08 | X-ray | LBO | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.53% | 22.70% | 6.20% | 2-(3-cyclopropylphenyl)-N-(4-methylpyridin-3-yl)acetamide | 2-(3-cyclopropylphenyl)-N-(4-methylpyridin-3-yl)acetamide | XDS | 99.40% | 11.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 58.0 | - | - | - | |||||
| 7gcd | 1.81 Å | 2023-11-08 | X-ray | LCU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.75% | 21.53% | 12.20% | 2-[(1R,3s,5S)-bicyclo[3.1.0]hexan-3-yl]-N-(4-methylpyridin-3-yl)acetamide | 2-[(1R,3s,5S)-bicyclo[3.1.0]hexan-3-yl]-N-(4-methylpyridin-3-yl)acetamide | XDS | 99.70% | 5.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 69.5 | - | - | - | |||||
| 7gce | 1.39 Å | 2023-11-08 | X-ray | LDX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.44% | 20.43% | 8.30% | 3-(2-fluorophenyl)-N-(4-methylpyridin-3-yl)propanamide | 3-(2-fluorophenyl)-N-(4-methylpyridin-3-yl)propanamide | XDS | 94.80% | 9.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 78.6 | - | - | - | |||||
| 7gcf | 1.42 Å | 2023-11-08 | X-ray | LKX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.16% | 21.25% | 5.90% | 2-(3-chlorophenyl)-N-(5-oxo-1,5-dihydro-4H-1,2,4-triazol-4-yl)acetamide | 2-(3-chlorophenyl)-N-(5-oxo-1,5-dihydro-4H-1,2,4-triazol-4-yl)acetamide | XDS | 92.80% | 9.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 72.0 | - | - | - | |||||
| 7gcg | 1.89 Å | 2023-11-08 | X-ray | LKI | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.90% | 24.57% | 11.70% | 3-chloro-N-(4-methylpyridin-3-yl)benzene-1-sulfonamide | 3-chloro-N-(4-methylpyridin-3-yl)benzene-1-sulfonamide | XDS | 99.40% | 8.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 39.5 | - | - | - | |||||
| 7gci | 1.54 Å | 2023-11-08 | X-ray | LJO | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.87% | 21.64% | 13.30% | (3R)-3-cyano-N-(4-methylpyridin-3-yl)oxolane-3-carboxamide | (3R)-3-cyano-N-(4-methylpyridin-3-yl)oxolane-3-carboxamide | XDS | 98.40% | 3.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 68.5 | - | - | - | |||||
| 7gcj | 1.34 Å | 2023-11-08 | X-ray | LJ0 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.98% | 20.41% | 7.00% | (1R,6S,7r)-N-(4-methylpyridin-3-yl)bicyclo[4.1.0]heptane-7-carboxamide | (1R,6S,7r)-N-(4-methylpyridin-3-yl)bicyclo[4.1.0]heptane-7-carboxamide | XDS | 90.00% | 9.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 78.8 | - | - | - | |||||
| 7gck | 1.40 Å | 2023-11-08 | X-ray | LF3 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.45% | 21.68% | 6.50% | 2-(6-chloro-3-oxo-2,3-dihydro-4H-1,4-benzoxazin-4-yl)-N-(4-methylpyridin-3-yl)acetamide | 2-(6-chloro-3-oxo-2,3-dihydro-4H-1,4-benzoxazin-4-yl)-N-(4-methylpyridin-3-yl)acetamide | XDS | 97.90% | 10.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 68.2 | - | - | - | |||||
| 7gcl | 1.83 Å | 2023-11-08 | X-ray | LR0 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.56% | 22.91% | 9.80% | (7R)-N-(4-acetamidopyridin-3-yl)-4-fluorobicyclo[4.2.0]octa-1,3,5-triene-7-carboxamide | (7R)-N-(4-acetamidopyridin-3-yl)-4-fluorobicyclo[4.2.0]octa-1,3,5-triene-7-carboxamide | XDS | 99.80% | 7.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 55.8 | - | - | - | |||||
| 7gcm | 1.57 Å | 2023-11-08 | X-ray | LQU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.04% | 22.70% | 4.10% | 3-[(4R)-2-acetyl-1,2,3,4-tetrahydroisoquinolin-4-yl]benzonitrile | 3-[(4R)-2-acetyl-1,2,3,4-tetrahydroisoquinolin-4-yl]benzonitrile | XDS | 79.10% | 11.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 58.0 | - | - | - | |||||
| 7gcn | 1.62 Å | 2023-11-08 | X-ray | LQ0 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.23% | 21.60% | 10.20% | 2-(6-chloro-1H-indol-1-yl)-N-(4-methylpyridin-3-yl)acetamide | 2-(6-chloro-1H-indol-1-yl)-N-(4-methylpyridin-3-yl)acetamide | XDS | 99.30% | 5.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 68.8 | - | - | - | |||||
| 7gco | 1.59 Å | 2023-11-08 | X-ray | LO0 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.64% | 21.68% | 10.70% | N-[2-(2-methoxyphenoxy)ethyl]-2-oxo-2,3-dihydropyridine-4-carboxamide | N-[2-(2-methoxyphenoxy)ethyl]-2-oxo-2,3-dihydropyridine-4-carboxamide | XDS | 99.60% | 6.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 68.2 | - | - | - | |||||
| 7gcp | 1.56 Å | 2023-11-08 | X-ray | LM0 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.46% | 21.43% | 7.50% | (3S)-N',2-diacetyl-1,2,3,4-tetrahydroisoquinoline-3-carbohydrazide | (3S)-N',2-diacetyl-1,2,3,4-tetrahydroisoquinoline-3-carbohydrazide | XDS | 99.10% | 9.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 70.4 | - | - | - | |||||
| 7gcq | 1.69 Å | 2023-11-08 | X-ray | LRC | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.84% | 23.87% | 8.20% | 2-(5-chloropyridin-3-yl)-N-(4-methylpyridin-3-yl)acetamide | 2-(5-chloropyridin-3-yl)-N-(4-methylpyridin-3-yl)acetamide | XDS | 96.80% | 8.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 46.2 | - | - | - | |||||
| 7gcr | 1.67 Å | 2023-11-08 | X-ray | LRN | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.03% | 22.39% | 6.10% | 1-[(3R)-3-{(cyclohexylmethyl)[(1r,4R)-4-hydroxycyclohexyl]amino}piperidin-1-yl]ethan-1-one | 1-[(3R)-3-{(cyclohexylmethyl)[(1r,4R)-4-hydroxycyclohexyl]amino}piperidin-1-yl]ethan-1-one | XDS | 99.60% | 12.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 61.4 | - | - | - | |||||
| 7gcs | 1.62 Å | 2023-11-08 | X-ray | LS0 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.65% | 22.29% | 8.70% | 2-(6-fluoro-1H-indol-1-yl)-N-(4-methylpyridin-3-yl)acetamide | 2-(6-fluoro-1H-indol-1-yl)-N-(4-methylpyridin-3-yl)acetamide | XDS | 99.30% | 7.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 62.4 | - | - | - | |||||
| 7gct | 1.86 Å | 2023-11-08 | X-ray | LSF | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.78% | 23.52% | 6.20% | N-(4-methylpyridin-3-yl)-2-(3-{[(2R)-4-oxoazetidin-2-yl]oxy}phenyl)acetamide | N-(4-methylpyridin-3-yl)-2-(3-{[(2R)-4-oxoazetidin-2-yl]oxy}phenyl)acetamide | XDS | 96.30% | 10.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 49.7 | - | - | - | |||||
| 7gcu | 1.54 Å | 2023-11-08 | X-ray | LT9 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.21% | 20.80% | 9.10% | 1-(3-chlorophenyl)-N-(4-methylpyridin-3-yl)-3-oxocyclobutane-1-carboxamide | 1-(3-chlorophenyl)-N-(4-methylpyridin-3-yl)-3-oxocyclobutane-1-carboxamide | XDS | 98.40% | 7.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 75.6 | - | - | - | |||||
| 7gcv | 1.96 Å | 2023-11-08 | X-ray | LUC | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.67% | 22.89% | 7.40% | 2-(3-chloro-5-{[(2R)-4-oxoazetidin-2-yl]oxy}phenyl)-N-(4-methylpyridin-3-yl)acetamide | 2-(3-chloro-5-{[(2R)-4-oxoazetidin-2-yl]oxy}phenyl)-N-(4-methylpyridin-3-yl)acetamide | XDS | 98.20% | 10.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 56.3 | - | - | - | |||||
| 7gcw | 1.58 Å | 2023-11-08 | X-ray | LV9 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.25% | 21.11% | 6.40% | N-(4-benzyloxan-4-yl)-N'-(pyridin-3-yl)urea | N-(4-benzyloxan-4-yl)-N'-(pyridin-3-yl)urea | XDS | 99.60% | 11.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 73.1 | - | - | - | |||||
| 7gcx | 1.75 Å | 2023-11-08 | X-ray | LVM | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.09% | 22.52% | 14.10% | N-(4-methylpyridin-3-yl)-2-(spiro[2.3]hexan-5-yl)acetamide | N-(4-methylpyridin-3-yl)-2-(spiro[2.3]hexan-5-yl)acetamide | XDS | 96.80% | 7.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 60.0 | - | - | - | |||||
| 7gcy | 1.62 Å | 2023-11-08 | X-ray | LW6 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.30% | 21.68% | 8.80% | 1H-indole-4-carbaldehyde | 1H-indole-4-carbaldehyde | XDS | 99.00% | 7.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 68.2 | - | - | - | |||||
| 7gcz | 1.51 Å | 2023-11-08 | X-ray | LWO | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.33% | 22.78% | 8.20% | 2-(1H-benzotriazol-1-yl)-N-[4-(methylamino)phenyl]-N-[(thiophen-3-yl)methyl]acetamide | 2-(1H-benzotriazol-1-yl)-N-[4-(methylamino)phenyl]-N-[(thiophen-3-yl)methyl]acetamide | XDS | 97.50% | 7.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 57.4 | - | - | - | |||||
| 7gd0 | 1.65 Å | 2023-11-08 | X-ray | M4L | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.01% | 22.97% | 7.10% | (2S)-2-(3-bromophenyl)-2-hydroxy-N-(4-methoxypyridin-3-yl)acetamide | (2S)-2-(3-bromophenyl)-2-hydroxy-N-(4-methoxypyridin-3-yl)acetamide | XDS | 99.60% | 8.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 55.4 | - | - | - | |||||
| 7gd1 | 1.52 Å | 2023-11-08 | X-ray | M2X | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.15% | 20.80% | 8.10% | (2R)-2-amino-2-(5-bromo-2-methoxyphenyl)-N-(4-methylpyridin-3-yl)acetamide | (2R)-2-amino-2-(5-bromo-2-methoxyphenyl)-N-(4-methylpyridin-3-yl)acetamide | XDS | 98.00% | 8.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 75.6 | - | - | - | |||||
| 7gd2 | 1.65 Å | 2023-11-08 | X-ray | M26 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.55% | 21.17% | 6.50% | N-[(1R)-1-(3-bromophenyl)-2-methoxyethyl]-2-[(3S)-5-fluoro-2-oxo-2,3-dihydro-1H-indol-3-yl]acetamide | N-[(1R)-1-(3-bromophenyl)-2-methoxyethyl]-2-[(3S)-5-fluoro-2-oxo-2,3-dihydro-1H-indol-3-yl]acetamide | XDS | 99.60% | 10.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 72.7 | - | - | - | |||||
| 7gd3 | 1.75 Å | 2023-11-08 | X-ray | Y6J | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.93% | 22.23% | 9.90% | ~{N}-[4-[2-(benzotriazol-1-yl)ethanoyl-(thiophen-3-ylmethyl)amino]phenyl]cyclopropanecarboxamide | ~{N}-[4-[2-(benzotriazol-1-yl)ethanoyl-(thiophen-3-ylmethyl)amino]phenyl]cyclopropanecarboxamide | XDS | 99.60% | 7.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 62.8 | - | - | - | |||||
| 7gd4 | 1.75 Å | 2023-11-08 | X-ray | R30 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.86% | 22.17% | 7.80% | N-{4-[(1H-benzotriazol-1-ylacetyl)(thiophen-3-ylmethyl)amino]phenyl}propanamide | N-{4-[(1H-benzotriazol-1-ylacetyl)(thiophen-3-ylmethyl)amino]phenyl}propanamide | XDS | 99.40% | 9.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 63.5 | - | - | - | |||||
| 7gd5 | 1.81 Å | 2023-11-08 | X-ray | M0X | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.73% | 22.03% | 11.40% | 2-(1H-benzotriazol-1-yl)-N-[4-(dimethylamino)phenyl]-N-[(thiophen-3-yl)methyl]acetamide | 2-(1H-benzotriazol-1-yl)-N-[4-(dimethylamino)phenyl]-N-[(thiophen-3-yl)methyl]acetamide | XDS | 99.50% | 8.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 64.8 | - | - | - | |||||
| 7gd6 | 1.55 Å | 2023-11-08 | X-ray | M0G | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.61% | 21.05% | 6.90% | (2S)-2-(3-chlorophenyl)-2-hydroxy-N-(4-methylpyridin-3-yl)butanamide | (2S)-2-(3-chlorophenyl)-2-hydroxy-N-(4-methylpyridin-3-yl)butanamide | XDS | 99.00% | 8.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 73.7 | - | - | - | |||||
| 7gd7 | 1.51 Å | 2023-11-08 | X-ray | LZX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.87% | 21.11% | 7.00% | (2R)-4-[(methanesulfonyl)amino]-2-phenyl-N-(pyridin-3-yl)butanamide | (2R)-4-[(methanesulfonyl)amino]-2-phenyl-N-(pyridin-3-yl)butanamide | XDS | 98.90% | 9.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 73.1 | - | - | - | |||||
| 7gd8 | 1.64 Å | 2023-11-08 | X-ray | KU6 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.33% | 21.81% | 11.30% | (4R)-6-chloro-N-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 99.70% | 7.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 66.9 | - | - | - | |||||
| 7gd9 | 1.62 Å | 2023-11-08 | X-ray | LXF | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.35% | 21.16% | 7.10% | N-{4-[(2-phenylethyl)sulfamoyl]-1,3-benzothiazol-2-yl}acetamide | N-{4-[(2-phenylethyl)sulfamoyl]-1,3-benzothiazol-2-yl}acetamide | XDS | 99.20% | 9.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 72.7 | - | - | - | |||||
| 7gda | 1.71 Å | 2023-11-08 | X-ray | MF0 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.58% | 21.49% | 8.80% | (2R)-2-(5-chloropyridin-3-yl)-N-(4-methylpyridin-3-yl)propanamide | (2R)-2-(5-chloropyridin-3-yl)-N-(4-methylpyridin-3-yl)propanamide | XDS | 97.70% | 6.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 70.0 | - | - | - | |||||
| 7gdb | 1.70 Å | 2023-11-08 | X-ray | M9U | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.32% | 21.57% | 13.50% | (4S)-6-chloro-N-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4S)-6-chloro-N-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 99.30% | 6.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 69.2 | - | - | - | |||||
| 7gdc | 1.72 Å | 2023-11-08 | X-ray | M93 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.80% | 21.73% | 15.80% | (4R)-6-chloro-N-[4-(hydroxymethyl)pyridin-3-yl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-[4-(hydroxymethyl)pyridin-3-yl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 99.90% | 8.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 67.7 | - | - | - | |||||
| 7gdd | 1.34 Å | 2023-11-08 | X-ray | 860 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.92% | 20.14% | 6.60% | 2-(3-chlorophenyl)-N-(isoquinolin-4-yl)acetamide | 2-(3-chlorophenyl)-N-(isoquinolin-4-yl)acetamide | XDS | 93.70% | 10.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 80.7 | - | - | - | |||||
| 7gde | 1.50 Å | 2023-11-08 | X-ray | M7X | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.14% | 22.38% | 14.40% | 2-(3-chlorophenyl)-N-(5-methylpyridin-3-yl)acetamide | 2-(3-chlorophenyl)-N-(5-methylpyridin-3-yl)acetamide | XDS | 95.60% | 4.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 61.5 | - | - | - | |||||
| 7gdf | 1.96 Å | 2023-11-08 | X-ray | M6U | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.36% | 23.12% | 15.00% | N-(4-ethylpyridin-3-yl)-2-[6-(trifluoromethyl)pyridin-2-yl]acetamide | N-(4-ethylpyridin-3-yl)-2-[6-(trifluoromethyl)pyridin-2-yl]acetamide | XDS | 99.70% | 6.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 53.8 | - | - | - | |||||
| 7gdg | 1.81 Å | 2023-11-08 | X-ray | M6I | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.29% | 22.62% | 15.20% | N-(3-chlorophenyl)-N'-(pyridin-3-yl)urea | N-(3-chlorophenyl)-N'-(pyridin-3-yl)urea | XDS | 99.90% | 5.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 58.9 | - | - | - | |||||
| 7gdh | 1.89 Å | 2023-11-08 | X-ray | M5X | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.11% | 23.79% | 8.10% | (3S)-3-hydroxy-2-oxo-2,3-dihydro-1H-indole-5-sulfonamide | (3S)-3-hydroxy-2-oxo-2,3-dihydro-1H-indole-5-sulfonamide | XDS | 99.70% | 11.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 47.1 | - | - | - | |||||
| 7gdi | 1.80 Å | 2023-11-08 | X-ray | M5I | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.95% | 24.24% | 9.80% | N-(3-chlorophenyl)-2-(4-methylpyridin-3-yl)acetamide | N-(3-chlorophenyl)-2-(4-methylpyridin-3-yl)acetamide | XDS | 80.30% | 7.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 42.6 | - | - | - | |||||
| 7gdj | 1.77 Å | 2023-11-08 | X-ray | M50 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.78% | 26.78% | 21.00% | 2-(3-chlorophenyl)-N-(5-methylpyridazin-4-yl)acetamide | 2-(3-chlorophenyl)-N-(5-methylpyridazin-4-yl)acetamide | XDS | 93.80% | 4.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 21.1 | - | - | - | |||||
| 7gdk | 1.74 Å | 2023-11-08 | X-ray | MJR | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.26% | 23.22% | 13.40% | 2-(3-chlorophenyl)-N-(3-methyl-1H-pyrazol-4-yl)acetamide | 2-(3-chlorophenyl)-N-(3-methyl-1H-pyrazol-4-yl)acetamide | XDS | 93.70% | 6.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 52.8 | - | - | - | |||||
| 7gdl | 1.86 Å | 2023-11-08 | X-ray | MKI | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.19% | 23.12% | 21.10% | 2-(3-chlorophenyl)-N-methyl-N-(4-methylpyridin-3-yl)acetamide | 2-(3-chlorophenyl)-N-methyl-N-(4-methylpyridin-3-yl)acetamide | XDS | 98.70% | 5.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 53.8 | - | - | - | |||||
| 7gdm | 1.52 Å | 2023-11-08 | X-ray | MQ3 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.09% | 21.65% | 11.00% | (2R)-2-cyclohexyl-N-(4-methylpyridin-3-yl)propanamide | (2R)-2-cyclohexyl-N-(4-methylpyridin-3-yl)propanamide | XDS | 99.20% | 7.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 68.4 | - | - | - | |||||
| 7gdn | 1.35 Å | 2023-11-08 | X-ray | MU3 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 15.91% | 19.67% | 6.00% | (4R)-N-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-N-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 93.50% | 9.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 83.9 | - | - | - | |||||
| 7gdo | 1.76 Å | 2023-11-08 | X-ray | MVR | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.67% | 26.73% | 8.80% | N-[(1-methyl-1H-pyrazol-3-yl)methyl]-2-(pyridin-3-yl)-N-[4-(pyridin-2-yl)phenyl]acetamide | N-[(1-methyl-1H-pyrazol-3-yl)methyl]-2-(pyridin-3-yl)-N-[4-(pyridin-2-yl)phenyl]acetamide | XDS | 92.80% | 7.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 21.3 | - | - | - | |||||
| 7gdp | 1.82 Å | 2023-11-08 | X-ray | MVX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.55% | 23.86% | 10.30% | (3S)-5-chloro-N-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)-2,3-dihydro-1-benzofuran-3-carboxamide | (3S)-5-chloro-N-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)-2,3-dihydro-1-benzofuran-3-carboxamide | XDS | 99.60% | 8.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 46.3 | - | - | - | |||||
| 7gdq | 1.72 Å | 2023-11-08 | X-ray | MWN | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.40% | 21.21% | 11.20% | methyl (3R)-5-bromo-3-hydroxy-2-oxo-2,3-dihydro-1H-indole-7-carboxylate | methyl (3R)-5-bromo-3-hydroxy-2-oxo-2,3-dihydro-1H-indole-7-carboxylate | XDS | 99.00% | 6.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 72.2 | - | - | - | |||||
| 7gdr | 1.66 Å | 2023-11-08 | X-ray | MX9 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.31% | 23.32% | 7.80% | (3-methylphenyl)methyl (3R)-3-hydroxy-2-oxo-2,3-dihydro-1H-indole-7-carboxylate | (3-methylphenyl)methyl (3R)-3-hydroxy-2-oxo-2,3-dihydro-1H-indole-7-carboxylate | XDS | 99.00% | 9.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 51.8 | - | - | - | |||||
| 7gds | 1.46 Å | 2023-11-08 | X-ray | MZF | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.49% | 20.45% | 9.30% | (3R)-5-bromo-3-hydroxy-1-[(1,2,4-oxadiazol-3-yl)methyl]-1,3-dihydro-2H-indol-2-one | (3R)-5-bromo-3-hydroxy-1-[(1,2,4-oxadiazol-3-yl)methyl]-1,3-dihydro-2H-indol-2-one | XDS | 98.90% | 6.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 78.5 | - | - | - | |||||
| 7gdt | 1.51 Å | 2023-11-08 | X-ray | N00 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.83% | 21.57% | 8.10% | (2R)-2-(6-chloro-1-methyl-9H-carbazol-2-yl)propanoic acid | (2R)-2-(6-chloro-1-methyl-9H-carbazol-2-yl)propanoic acid | XDS | 96.10% | 8.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 69.2 | - | - | - | |||||
| 7gdu | 1.63 Å | 2023-11-08 | X-ray | N0F | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.49% | 21.36% | 9.80% | (3R)-3-(4-hydroxypiperidin-1-yl)-N-(4-methylpyridin-3-yl)-3-(thiophen-3-yl)propanamide | (3R)-3-(4-hydroxypiperidin-1-yl)-N-(4-methylpyridin-3-yl)-3-(thiophen-3-yl)propanamide | XDS | 99.10% | 7.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 71.0 | - | - | - | |||||
| 7gdv | 1.91 Å | 2023-11-08 | X-ray | N0O | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.44% | 24.12% | 10.80% | (3S)-5-chloro-N-(4-phenyl-4H-1,2,4-triazol-3-yl)-2,3-dihydro-1-benzofuran-3-carboxamide | (3S)-5-chloro-N-(4-phenyl-4H-1,2,4-triazol-3-yl)-2,3-dihydro-1-benzofuran-3-carboxamide | XDS | 99.40% | 7.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 43.7 | - | - | - | |||||
| 7gdw | 1.67 Å | 2023-11-08 | X-ray | N0X | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 15.95% | 22.13% | 8.40% | (4R)-N-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)-4-methyl-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-N-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)-4-methyl-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 90.00% | 8.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 63.8 | - | - | - | |||||
| 7gdx | 1.84 Å | 2023-11-08 | X-ray | N1U | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.61% | 22.89% | 12.50% | (3S)-5-bromo-1-[(3,4-dimethoxyphenyl)methyl]-3-hydroxy-7-methyl-1,3-dihydro-2H-indol-2-one | (3S)-5-bromo-1-[(3,4-dimethoxyphenyl)methyl]-3-hydroxy-7-methyl-1,3-dihydro-2H-indol-2-one | XDS | 99.80% | 6.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 56.3 | - | - | - | |||||
| 7gdy | 1.61 Å | 2023-11-08 | X-ray | N2X | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.59% | 25.86% | 10.90% | (3S)-5-bromo-1-[(2-ethoxyphenyl)methyl]-3-hydroxy-1,3-dihydro-2H-indol-2-one | (3S)-5-bromo-1-[(2-ethoxyphenyl)methyl]-3-hydroxy-1,3-dihydro-2H-indol-2-one | XDS | 97.50% | 5.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 27.8 | - | - | - | |||||
| 7gdz | 1.84 Å | 2023-11-08 | X-ray | N3I | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.60% | 24.57% | 9.00% | 2-(3-chloro-5-methoxyphenyl)-N-(4-methylpyridin-3-yl)acetamide | 2-(3-chloro-5-methoxyphenyl)-N-(4-methylpyridin-3-yl)acetamide | XDS | 97.70% | 7.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 39.5 | - | - | - | |||||
| 7ge0 | 1.70 Å | 2023-11-08 | X-ray | N3R | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.94% | 24.77% | 11.70% | 5-fluoro-N-[2-(2-methoxyphenoxy)ethyl]-2-oxo-1,2-dihydroquinoline-4-carboxamide | 5-fluoro-N-[2-(2-methoxyphenoxy)ethyl]-2-oxo-1,2-dihydroquinoline-4-carboxamide | XDS | 99.50% | 7.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 37.5 | - | - | - | |||||
| 7ge1 | 1.94 Å | 2023-11-08 | X-ray | N43 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.57% | 25.17% | 10.10% | 5-methoxy-N-[2-(2-methoxyphenoxy)ethyl]-2-oxo-1,2-dihydroquinoline-4-carboxamide | 5-methoxy-N-[2-(2-methoxyphenoxy)ethyl]-2-oxo-1,2-dihydroquinoline-4-carboxamide | XDS | 99.00% | 9.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 33.8 | - | - | - | |||||
| 7ge2 | 1.67 Å | 2023-11-08 | X-ray | N4L | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.65% | 24.04% | 11.40% | N-[2-(2-methoxyphenoxy)ethyl]-5-methyl-2-oxo-1,2-dihydroquinoline-4-carboxamide | N-[2-(2-methoxyphenoxy)ethyl]-5-methyl-2-oxo-1,2-dihydroquinoline-4-carboxamide | XDS | 99.20% | 7.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 44.5 | - | - | - | |||||
| 7ge3 | 1.27 Å | 2023-11-08 | X-ray | N5L | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.61% | 19.23% | 6.80% | N-(5-amino-4-methylpyridin-3-yl)-2-(3-chlorophenyl)acetamide | N-(5-amino-4-methylpyridin-3-yl)-2-(3-chlorophenyl)acetamide | XDS | 93.50% | 9.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 86.5 | - | - | - | |||||
| 7ge4 | 1.30 Å | 2023-11-08 | X-ray | N6X | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.76% | 20.02% | 7.60% | N-(5-amino-4-methylpyridin-3-yl)-2-(3-cyanophenyl)acetamide | N-(5-amino-4-methylpyridin-3-yl)-2-(3-cyanophenyl)acetamide | XDS | 93.50% | 7.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 81.5 | - | - | - | |||||
| 7ge5 | 1.64 Å | 2023-11-08 | X-ray | N7L | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.18% | 22.52% | 13.50% | (1R,2R)-2-(fluoromethyl)-N-(4-methylpyridin-3-yl)cyclopropane-1-carboxamide | (1R,2R)-2-(fluoromethyl)-N-(4-methylpyridin-3-yl)cyclopropane-1-carboxamide | XDS | 99.30% | 5.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 60.0 | - | - | - | |||||
| 7ge6 | 1.62 Å | 2023-11-08 | X-ray | N8X | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.95% | 21.80% | 9.40% | 2-(3,5-dimethylphenyl)-N-(4-methyl-4H-1,2,4-triazol-3-yl)acetamide | 2-(3,5-dimethylphenyl)-N-(4-methyl-4H-1,2,4-triazol-3-yl)acetamide | XDS | 99.80% | 9.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 66.9 | - | - | - | |||||
| 7ge7 | 1.51 Å | 2023-11-08 | X-ray | N9I | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.61% | 20.69% | 9.40% | 2-(4-methylpyridin-3-yl)-N-(1,2,3,4-tetrahydroisoquinolin-8-yl)acetamide | 2-(4-methylpyridin-3-yl)-N-(1,2,3,4-tetrahydroisoquinolin-8-yl)acetamide | XDS | 99.70% | 7.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 76.7 | - | - | - | |||||
| 7ge8 | 1.80 Å | 2023-11-08 | X-ray | NB0 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.67% | 22.47% | 7.70% | 4-[4-(2-fluorophenyl)piperazine-1-carbonyl]quinolin-2(1H)-one | 4-[4-(2-fluorophenyl)piperazine-1-carbonyl]quinolin-2(1H)-one | XDS | 99.60% | 6.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 60.6 | - | - | - | |||||
| 7ge9 | 1.49 Å | 2023-11-08 | X-ray | NB6 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.80% | 20.57% | 9.60% | 2-(3-bromophenyl)-N-(4-methylpyridin-3-yl)acetamide | 2-(3-bromophenyl)-N-(4-methylpyridin-3-yl)acetamide | XDS | 99.70% | 9.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 77.6 | - | - | - | |||||
| 7gea | 1.83 Å | 2023-11-08 | X-ray | N6F | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.68% | 22.86% | 9.70% | (2S)-N-(4-acetamidopyridin-3-yl)-2-(3-chlorophenyl)propanamide | (2S)-N-(4-acetamidopyridin-3-yl)-2-(3-chlorophenyl)propanamide | XDS | 99.90% | 8.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 56.5 | - | - | - | |||||
| 7geb | 1.39 Å | 2023-11-08 | X-ray | NDI | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.58% | 21.90% | 7.70% | 2-(4-acetylpiperazin-1-yl)-N-(4-methylpyridin-3-yl)acetamide | 2-(4-acetylpiperazin-1-yl)-N-(4-methylpyridin-3-yl)acetamide | XDS | 98.60% | 6.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 65.9 | - | - | - | |||||
| 7gec | 1.25 Å | 2023-11-08 | X-ray | NEL | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.88% | 19.70% | 6.90% | 2-(3-chlorophenyl)-N-(1H-indazol-4-yl)acetamide | 2-(3-chlorophenyl)-N-(1H-indazol-4-yl)acetamide | XDS | 92.80% | 7.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 83.7 | - | - | - | |||||
| 7ged | 1.48 Å | 2023-11-08 | X-ray | NGX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.77% | 22.28% | 10.40% | (2S)-1-(3-chlorophenyl)-3-(1H-1,2,4-triazol-1-yl)propan-2-ol | (2S)-1-(3-chlorophenyl)-3-(1H-1,2,4-triazol-1-yl)propan-2-ol | XDS | 99.20% | 7.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 62.5 | - | - | - | |||||
| 7gee | 1.29 Å | 2023-11-08 | X-ray | NIJ | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.03% | 19.77% | 6.20% | 3-(3-fluorophenyl)-N-(4-methylpyridin-3-yl)propanamide | 3-(3-fluorophenyl)-N-(4-methylpyridin-3-yl)propanamide | XDS | 86.00% | 11.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 83.3 | - | - | - | |||||
| 7gef | 1.18 Å | 2023-11-08 | X-ray | NJE | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.76% | 18.65% | 6.60% | N-(4-methylpyridin-3-yl)-2-(piperidin-1-yl)acetamide | N-(4-methylpyridin-3-yl)-2-(piperidin-1-yl)acetamide | XDS | 81.80% | 11.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 89.5 | - | - | - | |||||
| 7geg | 1.73 Å | 2023-11-08 | X-ray | NJU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.83% | 22.23% | 10.30% | 2-(3-chlorophenyl)-N-(5,6,7,8-tetrahydroisoquinolin-4-yl)acetamide | 2-(3-chlorophenyl)-N-(5,6,7,8-tetrahydroisoquinolin-4-yl)acetamide | XDS | 99.70% | 7.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 62.8 | - | - | - | |||||
| 7geh | 1.23 Å | 2023-11-08 | X-ray | NKU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.79% | 19.66% | 6.90% | 2-(1H-benzotriazol-1-yl)-N-[(3-chlorophenyl)methyl]-N-methylacetamide | 2-(1H-benzotriazol-1-yl)-N-[(3-chlorophenyl)methyl]-N-methylacetamide | XDS | 89.20% | 9.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 84.0 | - | - | - | |||||
| 7gei | 1.69 Å | 2023-11-08 | X-ray | NM0 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.43% | 23.15% | 8.60% | 2-(3-chlorophenyl)-N-[(4R)-imidazo[1,2-a]pyridin-3-yl]acetamide | 2-(3-chlorophenyl)-N-[(4R)-imidazo[1,2-a]pyridin-3-yl]acetamide | XDS | 99.90% | 12.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 53.6 | - | - | - | |||||
| 7gej | 1.74 Å | 2023-11-08 | X-ray | NO0 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.35% | 25.12% | 14.60% | 2-(3-ethynylphenyl)-N-(isoquinolin-4-yl)acetamide | 2-(3-ethynylphenyl)-N-(isoquinolin-4-yl)acetamide | XDS | 99.80% | 10.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 34.0 | - | - | - | |||||
| 7gek | 1.41 Å | 2023-11-08 | X-ray | NOI | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.32% | 21.42% | 7.90% | 2-(3-chlorophenyl)-N-(1-methyl-1H-imidazol-5-yl)acetamide | 2-(3-chlorophenyl)-N-(1-methyl-1H-imidazol-5-yl)acetamide | XDS | 97.70% | 13.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 70.4 | - | - | - | |||||
| 7gel | 1.49 Å | 2023-11-08 | X-ray | NQ3 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.71% | 21.71% | 10.50% | (1M,3P)-1-(3-chlorophenyl)-3-(4-methylpyridin-3-yl)-1,3-dihydro-2H-imidazol-2-one | (1M,3P)-1-(3-chlorophenyl)-3-(4-methylpyridin-3-yl)-1,3-dihydro-2H-imidazol-2-one | XDS | 99.70% | 12.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 67.8 | - | - | - | |||||
| 7gem | 1.32 Å | 2023-11-08 | X-ray | NQO | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.76% | 20.64% | 8.00% | 2-(3-chlorophenyl)-N-(4-methylpyridazin-3-yl)acetamide | 2-(3-chlorophenyl)-N-(4-methylpyridazin-3-yl)acetamide | XDS | 98.00% | 12.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 77.0 | - | - | - | |||||
| 7gen | 1.53 Å | 2023-11-08 | X-ray | NRC | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.26% | 21.01% | 10.30% | methyl (2R)-2-(3-chlorophenyl)-3-[(4-methylpyridin-3-yl)amino]-3-oxopropanoate | methyl (2R)-2-(3-chlorophenyl)-3-[(4-methylpyridin-3-yl)amino]-3-oxopropanoate | XDS | 99.90% | 12.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 73.9 | - | - | - | |||||
| 7geo | 1.50 Å | 2023-11-08 | X-ray | NRX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.81% | 22.43% | 9.50% | 2-(3-chlorophenyl)-N-[(4S)-imidazo[1,5-a]pyridin-1-yl]acetamide | 2-(3-chlorophenyl)-N-[(4S)-imidazo[1,5-a]pyridin-1-yl]acetamide | XDS | 99.50% | 8.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 60.9 | - | - | - | |||||
| 7geq | 2.06 Å | 2023-11-08 | X-ray | NSR | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.20% | 25.74% | 22.50% | (4R)-6-chloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 99.50% | 15.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 28.8 | - | - | - | |||||
| 7ger | 1.71 Å | 2023-11-08 | X-ray | NU0 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.54% | 28.03% | 20.20% | 2-(3-chlorophenyl)-N-(2,6-naphthyridin-4-yl)acetamide | 2-(3-chlorophenyl)-N-(2,6-naphthyridin-4-yl)acetamide | XDS | 99.10% | 5.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 13.9 | - | - | - | |||||
| 7ges | 1.47 Å | 2023-11-08 | X-ray | NUR | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 16.95% | 21.33% | 11.40% | 2-(3-chlorophenyl)-N-(isoquinolin-4-yl)-N-methylacetamide | 2-(3-chlorophenyl)-N-(isoquinolin-4-yl)-N-methylacetamide | XDS | 99.40% | 10.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 71.2 | - | - | - | |||||
| 7get | 1.92 Å | 2023-11-08 | X-ray | NV9 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.76% | 25.40% | 9.00% | 2-(3-chlorophenyl)-N-(4-phenylpyridin-3-yl)acetamide | 2-(3-chlorophenyl)-N-(4-phenylpyridin-3-yl)acetamide | XDS | 99.60% | 13.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 31.6 | - | - | - | |||||
| 7geu | 1.40 Å | 2023-11-08 | X-ray | NVO | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.52% | 21.06% | 8.70% | 2-(3-chlorophenyl)-N-(phthalazin-1-yl)acetamide | 2-(3-chlorophenyl)-N-(phthalazin-1-yl)acetamide | XDS | 94.50% | 10.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 73.6 | - | - | - | |||||
| 7gev | 1.54 Å | 2023-11-08 | X-ray | NW0 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.90% | 21.52% | 9.10% | 3-(3-chlorophenyl)-N-(4-methylpyridin-3-yl)propanamide | 3-(3-chlorophenyl)-N-(4-methylpyridin-3-yl)propanamide | XDS | 99.40% | 12.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 69.6 | - | - | - | |||||
| 7gew | 1.46 Å | 2023-11-08 | X-ray | NWI | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.06% | 21.88% | 10.10% | 2-(3-chlorophenyl)-N-(1,6-naphthyridin-8-yl)acetamide | 2-(3-chlorophenyl)-N-(1,6-naphthyridin-8-yl)acetamide | XDS | 98.90% | 9.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 66.3 | - | - | - | |||||
| 7gex | 1.99 Å | 2023-11-08 | X-ray | NX9 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.55% | 21.64% | 14.30% | 2-(3-chlorophenyl)-N-(1H-pyrazol-4-yl)acetamide | 2-(3-chlorophenyl)-N-(1H-pyrazol-4-yl)acetamide | XDS | 99.60% | 18.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 68.5 | - | - | - | |||||
| 7gey | 1.34 Å | 2023-11-08 | X-ray | NYR | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.28% | 21.62% | 7.80% | N-(3-chlorophenyl)-2-(3-methyl-1H-pyrazol-4-yl)acetamide | N-(3-chlorophenyl)-2-(3-methyl-1H-pyrazol-4-yl)acetamide | XDS | 96.50% | 10.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 68.7 | - | - | - | |||||
| 7gez | 1.34 Å | 2023-11-08 | X-ray | NZK | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.44% | 20.41% | 6.60% | 2-(4-acetylpiperazin-1-yl)-N-(isoquinolin-4-yl)acetamide | 2-(4-acetylpiperazin-1-yl)-N-(isoquinolin-4-yl)acetamide | XDS | 97.10% | 12.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 78.8 | - | - | - | |||||
| 7gf0 | 1.58 Å | 2023-11-08 | X-ray | O0C | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.28% | 24.21% | 7.90% | 2-(3-chlorophenyl)-N-(2,7-naphthyridin-4-yl)acetamide | 2-(3-chlorophenyl)-N-(2,7-naphthyridin-4-yl)acetamide | XDS | 98.40% | 12.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 42.8 | - | - | - | |||||
| 7gf1 | 1.88 Å | 2023-11-08 | X-ray | O0R | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.39% | 25.48% | 14.00% | 2-(3-chlorophenyl)-N-(4-cyclopropylpyridin-3-yl)acetamide | 2-(3-chlorophenyl)-N-(4-cyclopropylpyridin-3-yl)acetamide | XDS | 99.80% | 8.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 31.1 | - | - | - | |||||
| 7gf2 | 1.38 Å | 2023-11-08 | X-ray | O0X | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.69% | 21.94% | 9.00% | 2-(3-chlorophenyl)-N-(1,7-naphthyridin-5-yl)acetamide | 2-(3-chlorophenyl)-N-(1,7-naphthyridin-5-yl)acetamide | XDS | 98.70% | 9.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 65.6 | - | - | - | |||||
| 7gf3 | 1.63 Å | 2023-11-08 | X-ray | O1I | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.02% | 25.97% | 10.80% | (2S)-2-(3-chlorophenyl)-3-hydroxy-N-(4-methylpyridin-3-yl)propanamide | (2S)-2-(3-chlorophenyl)-3-hydroxy-N-(4-methylpyridin-3-yl)propanamide | XDS | 99.10% | 11.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 26.9 | - | - | - | |||||
| 7gf4 | 1.56 Å | 2023-11-08 | X-ray | O1X | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.92% | 23.00% | 9.30% | (2S)-2-(difluoromethoxy)-N-(4-methylpyridin-3-yl)propanamide | (2S)-2-(difluoromethoxy)-N-(4-methylpyridin-3-yl)propanamide | XDS | 99.40% | 12.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 54.9 | - | - | - | |||||
| 7gf5 | 1.37 Å | 2023-11-08 | X-ray | O2R | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.05% | 21.27% | 6.80% | 2-(2-butoxy-5-chlorophenyl)-N-(4-methylpyridin-3-yl)acetamide | 2-(2-butoxy-5-chlorophenyl)-N-(4-methylpyridin-3-yl)acetamide | XDS | 98.30% | 11.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 71.8 | - | - | - | |||||
| 7gf6 | 1.45 Å | 2023-11-08 | X-ray | O3I | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.29% | 21.92% | 7.00% | 2-[(1M)-5-chloro-2',3'-difluoro-4'-methyl[1,1'-biphenyl]-3-yl]-N-(4-methylpyridin-3-yl)acetamide | 2-[(1M)-5-chloro-2',3'-difluoro-4'-methyl[1,1'-biphenyl]-3-yl]-N-(4-methylpyridin-3-yl)acetamide | XDS | 99.50% | 13.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 65.8 | - | - | - | |||||
| 7gf7 | 1.94 Å | 2023-11-08 | X-ray | O3U | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.93% | 24.10% | 24.50% | N-[(1R)-1,5-dicyano-4-(methylsulfanyl)-3-azaspiro[5.5]undeca-2,4-dien-2-yl]acetamide | N-[(1R)-1,5-dicyano-4-(methylsulfanyl)-3-azaspiro[5.5]undeca-2,4-dien-2-yl]acetamide | XDS | 99.70% | 5.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 43.8 | - | - | - | |||||
| 7gf8 | 1.43 Å | 2023-11-08 | X-ray | O4L | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.69% | 20.96% | 10.60% | N-(3-acetyl-2,5-dimethyl-1H-pyrrol-1-yl)-4-oxo-3,4-dihydrophthalazine-1-carboxamide | N-(3-acetyl-2,5-dimethyl-1H-pyrrol-1-yl)-4-oxo-3,4-dihydrophthalazine-1-carboxamide | XDS | 97.20% | 10.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 74.4 | - | - | - | |||||
| 7gf9 | 1.97 Å | 2023-11-08 | X-ray | O5C | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.78% | 24.04% | 13.80% | 1-[(4S)-3-(4-fluorobenzoyl)-2-methylindolizin-1-yl]ethan-1-one | 1-[(4S)-3-(4-fluorobenzoyl)-2-methylindolizin-1-yl]ethan-1-one | XDS | 99.80% | 4.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 44.5 | - | - | - | |||||
| 7gfa | 1.97 Å | 2023-11-08 | X-ray | Z26 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.75% | 26.28% | 28.30% | 2-(5-chloro-2-methoxyphenyl)-N-(isoquinolin-4-yl)acetamide | 2-(5-chloro-2-methoxyphenyl)-N-(isoquinolin-4-yl)acetamide | XDS | 99.90% | 2.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 24.7 | - | - | - | |||||
| 7gfb | 1.61 Å | 2023-11-08 | X-ray | NSR | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.00% | 22.44% | 11.90% | (4R)-6-chloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 99.70% | 5.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 60.8 | - | - | - | |||||
| 7gfc | 2.23 Å | 2023-11-08 | X-ray | O87 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.51% | 25.32% | 22.50% | 4-{4-[3-(trifluoromethyl)phenyl]piperazine-1-carbonyl}quinolin-2(1H)-one | 4-{4-[3-(trifluoromethyl)phenyl]piperazine-1-carbonyl}quinolin-2(1H)-one | XDS | 99.90% | 5.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 32.3 | - | - | - | |||||
| 7gfd | 1.85 Å | 2023-11-08 | X-ray | O8L | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.37% | 23.56% | 7.70% | (2S)-2-(3-chloro-5-{[(2R)-4-oxoazetidin-2-yl]oxy}phenyl)-N-(4-methylpyridin-3-yl)propanamide | (2S)-2-(3-chloro-5-{[(2R)-4-oxoazetidin-2-yl]oxy}phenyl)-N-(4-methylpyridin-3-yl)propanamide | XDS | 99.60% | 8.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 49.4 | - | - | - | |||||
| 7gfe | 1.65 Å | 2023-11-08 | X-ray | O9O | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.62% | 21.78% | 12.30% | N-{3-chloro-5-[(6-methoxypyridin-2-yl)oxy]phenyl}-2-(isoquinolin-4-yl)acetamide | N-{3-chloro-5-[(6-methoxypyridin-2-yl)oxy]phenyl}-2-(isoquinolin-4-yl)acetamide | XDS | 99.80% | 12.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 67.3 | - | - | - | |||||
| 7gff | 1.75 Å | 2023-11-08 | X-ray | O9X | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.52% | 22.27% | 11.30% | (2R)-2-(difluoromethoxy)-N-(4-methylpyridin-3-yl)propanamide | (2R)-2-(difluoromethoxy)-N-(4-methylpyridin-3-yl)propanamide | XDS | 99.40% | 13.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 62.5 | - | - | - | |||||
| 7gfg | 1.80 Å | 2023-11-08 | X-ray | OAO | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.31% | 25.01% | 9.10% | 2-{3-chloro-5-[(3-methyl-1,2,4-oxadiazol-5-yl)methoxy]phenyl}-N-(4-methylpyridin-3-yl)acetamide | 2-{3-chloro-5-[(3-methyl-1,2,4-oxadiazol-5-yl)methoxy]phenyl}-N-(4-methylpyridin-3-yl)acetamide | XDS | 99.30% | 6.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 35.0 | - | - | - | |||||
| 7gfh | 1.46 Å | 2023-11-08 | X-ray | OBO | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.97% | 22.26% | 8.90% | 2-(3-chlorophenyl)-N-(1H-imidazo[4,5-c]pyridin-7-yl)acetamide | 2-(3-chlorophenyl)-N-(1H-imidazo[4,5-c]pyridin-7-yl)acetamide | XDS | 96.00% | 12.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 62.6 | - | - | - | |||||
| 7gfi | 1.60 Å | 2023-11-08 | X-ray | OCI | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.48% | 23.26% | 7.30% | 2-(3-chloro-5-sulfamamidophenyl)-N-(4-methylpyridin-3-yl)acetamide | 2-(3-chloro-5-sulfamamidophenyl)-N-(4-methylpyridin-3-yl)acetamide | XDS | 99.20% | 13.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 52.5 | - | - | - | |||||
| 7gfj | 1.59 Å | 2023-11-08 | X-ray | OD7 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.99% | 22.18% | 7.70% | 2-(6-methoxy-1H-benzotriazol-1-yl)-N-[4-(piperidin-4-yl)phenyl]-N-[(pyridin-2-yl)methyl]acetamide | 2-(6-methoxy-1H-benzotriazol-1-yl)-N-[4-(piperidin-4-yl)phenyl]-N-[(pyridin-2-yl)methyl]acetamide | XDS | 99.80% | 12.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 63.4 | - | - | - | |||||
| 7gfk | 1.50 Å | 2023-11-08 | X-ray | ODX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.13% | 21.74% | 7.20% | 2-(3-chloro-5-{[(1S,2S)-2-hydroxycyclopentyl]amino}phenyl)-N-(4-methylpyridin-3-yl)acetamide | 2-(3-chloro-5-{[(1S,2S)-2-hydroxycyclopentyl]amino}phenyl)-N-(4-methylpyridin-3-yl)acetamide | XDS | 99.60% | 12.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 67.6 | - | - | - | |||||
| 7gfl | 1.77 Å | 2023-11-08 | X-ray | OE6 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.71% | 24.45% | 8.90% | N-(1H-benzotriazol-1-yl)-2-(3-chlorophenyl)acetamide | N-(1H-benzotriazol-1-yl)-2-(3-chlorophenyl)acetamide | XDS | 99.80% | 12.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 40.6 | - | - | - | |||||
| 7gfm | 1.70 Å | 2023-11-08 | X-ray | OEO | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.33% | 25.11% | 23.40% | N-[4-(dimethylamino)phenyl]-2-(isoquinolin-4-yl)-N-[(thiophen-3-yl)methyl]acetamide | N-[4-(dimethylamino)phenyl]-2-(isoquinolin-4-yl)-N-[(thiophen-3-yl)methyl]acetamide | XDS | 99.40% | 4.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 34.1 | - | - | - | |||||
| 7gfn | 1.64 Å | 2023-11-08 | X-ray | OFX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.24% | 23.58% | 12.60% | 2-(1H-benzotriazol-1-yl)-N-[4-(dimethylamino)phenyl]-N-[(1H-pyrazol-5-yl)methyl]acetamide | 2-(1H-benzotriazol-1-yl)-N-[4-(dimethylamino)phenyl]-N-[(1H-pyrazol-5-yl)methyl]acetamide | XDS | 99.80% | 11.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 49.2 | - | - | - | |||||
| 7gfo | 1.50 Å | 2023-11-08 | X-ray | OGF | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.88% | 22.30% | 12.70% | 2-(1H-benzotriazol-1-yl)-N-[4-(methylcarbamamido)phenyl]-N-[(thiophen-3-yl)methyl]acetamide | 2-(1H-benzotriazol-1-yl)-N-[4-(methylcarbamamido)phenyl]-N-[(thiophen-3-yl)methyl]acetamide | XDS | 99.20% | 5.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 62.1 | - | - | - | |||||
| 7gfp | 1.77 Å | 2023-11-08 | X-ray | OHC | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.19% | 24.35% | 13.20% | 2-[3-(acetamidomethyl)-5-chlorophenyl]-N-(4-methylpyridin-3-yl)acetamide | 2-[3-(acetamidomethyl)-5-chlorophenyl]-N-(4-methylpyridin-3-yl)acetamide | XDS | 99.80% | 6.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 41.6 | - | - | - | |||||
| 7gfq | 1.55 Å | 2023-11-08 | X-ray | OI4 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.58% | 22.79% | 13.20% | (4R)-6-chloro-N-(4-methylpyridin-3-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-(4-methylpyridin-3-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 98.50% | 8.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 57.3 | - | - | - | |||||
| 7gfr | 1.46 Å | 2023-11-08 | X-ray | OIE | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.86% | 22.11% | 13.20% | 2-(4-chloropyridin-2-yl)-N-(isoquinolin-4-yl)acetamide | 2-(4-chloropyridin-2-yl)-N-(isoquinolin-4-yl)acetamide | XDS | 99.70% | 9.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 64.0 | - | - | - | |||||
| 7gfs | 1.68 Å | 2023-11-08 | X-ray | OIK | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.09% | 23.38% | 6.30% | (isoquinolin-4-yl)(4-phenylpiperazin-1-yl)methanone | (isoquinolin-4-yl)(4-phenylpiperazin-1-yl)methanone | XDS | 99.70% | 15.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 51.3 | - | - | - | |||||
| 7gft | 1.64 Å | 2023-11-08 | X-ray | OIX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.31% | 23.58% | 14.20% | N-[2-(2-methoxyphenoxy)ethyl]isoquinoline-4-carboxamide | N-[2-(2-methoxyphenoxy)ethyl]isoquinoline-4-carboxamide | XDS | 99.70% | 6.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 49.2 | - | - | - | |||||
| 7gfu | 1.60 Å | 2023-11-08 | X-ray | OJ9 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.14% | 22.81% | 13.00% | 2-(5-chloro-1-benzofuran-7-yl)-N-(isoquinolin-4-yl)acetamide | 2-(5-chloro-1-benzofuran-7-yl)-N-(isoquinolin-4-yl)acetamide | XDS | 99.80% | 8.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 56.9 | - | - | - | |||||
| 7gfv | 1.73 Å | 2023-11-08 | X-ray | OJO | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.41% | 23.54% | 8.00% | 1-{4-[(3-chloro-5-hydroxyphenyl)methyl]piperazin-1-yl}ethan-1-one | 1-{4-[(3-chloro-5-hydroxyphenyl)methyl]piperazin-1-yl}ethan-1-one | XDS | 99.40% | 11.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 49.6 | - | - | - | |||||
| 7gfw | 1.57 Å | 2023-11-08 | X-ray | OK9 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.84% | 21.42% | 9.10% | N-{(1Z)-1-[5-(morpholin-4-yl)thiophen-2-yl]-3-oxoprop-1-en-2-yl}thiophene-2-carboxamide | N-{(1Z)-1-[5-(morpholin-4-yl)thiophen-2-yl]-3-oxoprop-1-en-2-yl}thiophene-2-carboxamide | XDS | 99.80% | 13.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 70.4 | - | - | - | |||||
| 7gfx | 1.55 Å | 2023-11-08 | X-ray | OKW | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.36% | 25.78% | 11.00% | 2-(3-chlorophenyl)-N-(6,7-dihydro-5H-cyclopenta[c]pyridin-4-yl)acetamide | 2-(3-chlorophenyl)-N-(6,7-dihydro-5H-cyclopenta[c]pyridin-4-yl)acetamide | XDS | 99.50% | 6.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 28.5 | - | - | - | |||||
| 7gfy | 1.55 Å | 2023-11-08 | X-ray | OLX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.39% | 21.45% | 9.40% | [(3R)-5-ethyl-3-hydroxy-2-oxo-2,3-dihydro-1H-indol-1-yl]acetic acid | [(3R)-5-ethyl-3-hydroxy-2-oxo-2,3-dihydro-1H-indol-1-yl]acetic acid | XDS | 99.50% | 11.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 70.2 | - | - | - | |||||
| 7gfz | 1.43 Å | 2023-11-08 | X-ray | ONU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.07% | 21.49% | 7.90% | 4-[3-(2-methoxyanilino)azetidine-1-carbonyl]quinolin-2(1H)-one | 4-[3-(2-methoxyanilino)azetidine-1-carbonyl]quinolin-2(1H)-one | XDS | 95.70% | 11.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 70.0 | - | - | - | |||||
| 7gg0 | 1.51 Å | 2023-11-08 | X-ray | OGV | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.97% | 22.82% | 8.40% | 2-(3-chlorophenyl)-N-[(4S)-[1,2,4]triazolo[4,3-a]pyridin-3-yl]acetamide | 2-(3-chlorophenyl)-N-[(4S)-[1,2,4]triazolo[4,3-a]pyridin-3-yl]acetamide | XDS | 98.00% | 10.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 56.8 | - | - | - | |||||
| 7gg1 | 1.48 Å | 2023-11-08 | X-ray | OGO | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.64% | 20.95% | 7.00% | 4-[3-(2-methoxyphenoxy)azetidine-1-carbonyl]quinolin-2(1H)-one | 4-[3-(2-methoxyphenoxy)azetidine-1-carbonyl]quinolin-2(1H)-one | XDS | 98.50% | 15.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 74.5 | - | - | - | |||||
| 7gg2 | 1.72 Å | 2023-11-08 | X-ray | OO6 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.01% | 27.49% | 21.00% | (3S,4R)-6-chloro-N-(isoquinolin-4-yl)-3-methyl-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (3S,4R)-6-chloro-N-(isoquinolin-4-yl)-3-methyl-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 99.90% | 4.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 16.8 | - | - | - | |||||
| 7gg3 | 1.42 Å | 2023-11-08 | X-ray | OOL | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.80% | 21.49% | 9.90% | (E)-1-(4,6-dimethoxypyrimidin-2-yl)methanimine | (E)-1-(4,6-dimethoxypyrimidin-2-yl)methanimine | XDS | 98.90% | 9.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 70.0 | - | - | - | |||||
| 7gg4 | 1.57 Å | 2023-11-08 | X-ray | OPU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.12% | 25.92% | 11.20% | (4R)-6-chloro-N-[(4S)-7-methyl[1,2,4]triazolo[4,3-a]pyridin-8-yl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-[(4S)-7-methyl[1,2,4]triazolo[4,3-a]pyridin-8-yl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 97.60% | 9.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 27.2 | - | - | - | |||||
| 7gg5 | 1.51 Å | 2023-11-08 | X-ray | OQF | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.59% | 21.36% | 7.80% | 2-(1H-benzotriazol-1-yl)-N-[4-(dimethylamino)phenyl]-N-[(1,3-thiazol-4-yl)methyl]acetamide | 2-(1H-benzotriazol-1-yl)-N-[4-(dimethylamino)phenyl]-N-[(1,3-thiazol-4-yl)methyl]acetamide | XDS | 98.00% | 14.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 71.0 | - | - | - | |||||
| 7gg6 | 1.60 Å | 2023-11-08 | X-ray | OQL | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.35% | 24.51% | 7.70% | (4R)-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide | (4R)-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide | XDS | 99.50% | 13.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 39.9 | - | - | - | |||||
| 7gg7 | 1.51 Å | 2023-11-08 | X-ray | OQX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.45% | 20.74% | 7.80% | 2-(1H-benzotriazol-1-yl)-N-[(3-chlorophenyl)methyl]-N-(4-methoxyphenyl)acetamide | 2-(1H-benzotriazol-1-yl)-N-[(3-chlorophenyl)methyl]-N-(4-methoxyphenyl)acetamide | XDS | 99.70% | 14.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 76.2 | - | - | - | |||||
| 7gg8 | 1.58 Å | 2023-11-08 | X-ray | ORR | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.81% | 21.82% | 8.20% | 1-(5-amino-3,4-dihydro-1,7-naphthyridin-1(2H)-yl)-2-(3-chlorophenyl)ethan-1-one | 1-(5-amino-3,4-dihydro-1,7-naphthyridin-1(2H)-yl)-2-(3-chlorophenyl)ethan-1-one | XDS | 99.60% | 14.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 66.8 | - | - | - | |||||
| 7gg9 | 1.54 Å | 2023-11-08 | X-ray | SQ2 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.18% | 22.97% | 12.80% | 2-(1H-1,2,3-benzotriazol-1-yl)-1-(4-methylpiperidin-1-yl)ethan-1-one | 2-(1H-1,2,3-benzotriazol-1-yl)-1-(4-methylpiperidin-1-yl)ethan-1-one | XDS | 99.40% | 7.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 55.4 | - | - | - | |||||
| 7gga | 1.49 Å | 2023-11-08 | X-ray | OSI | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.29% | 20.40% | 6.30% | (4R)-6-chloro-N-(isoquinolin-4-yl)-4-methyl-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-(isoquinolin-4-yl)-4-methyl-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 99.70% | 14.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 78.8 | - | - | - | |||||
| 7ggb | 1.38 Å | 2023-11-08 | X-ray | OT6 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.33% | 19.88% | 6.50% | 2-{3-chloro-5-[(2-cyano-2-methylpropyl)amino]phenyl}-N-(4-methylpyridin-3-yl)acetamide | 2-{3-chloro-5-[(2-cyano-2-methylpropyl)amino]phenyl}-N-(4-methylpyridin-3-yl)acetamide | XDS | 99.10% | 14.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 82.6 | - | - | - | |||||
| 7ggc | 1.94 Å | 2023-11-08 | X-ray | OTV | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.75% | 28.56% | 14.40% | 2-(3-chloro-5-{[(1S,2R)-2-(trifluoromethyl)cyclopropyl]amino}phenyl)-N-(4-methylpyridin-3-yl)acetamide | 2-(3-chloro-5-{[(1S,2R)-2-(trifluoromethyl)cyclopropyl]amino}phenyl)-N-(4-methylpyridin-3-yl)acetamide | XDS | 95.60% | 5.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 11.7 | - | - | - | |||||
| 7ggd | 1.48 Å | 2023-11-08 | X-ray | OUF | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.44% | 20.42% | 8.30% | N-[(furan-2-yl)methyl]-N'-(2-methyl-1-oxo-1,2-dihydroisoquinolin-4-yl)-N-{3-[(propan-2-yl)oxy]propyl}urea | N-[(furan-2-yl)methyl]-N'-(2-methyl-1-oxo-1,2-dihydroisoquinolin-4-yl)-N-{3-[(propan-2-yl)oxy]propyl}urea | XDS | 99.00% | 10.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 78.7 | - | - | - | |||||
| 7gge | 1.63 Å | 2023-11-08 | X-ray | OV4 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.73% | 24.83% | 18.90% | 2-(1H-benzotriazol-1-yl)-N-benzyl-N-[4-(dimethylamino)phenyl]acetamide | 2-(1H-benzotriazol-1-yl)-N-benzyl-N-[4-(dimethylamino)phenyl]acetamide | XDS | 98.00% | 4.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 36.8 | - | - | - | |||||
| 7ggf | 1.83 Å | 2023-11-08 | X-ray | OVF | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.91% | 28.17% | 17.80% | 2-(1H-benzotriazol-1-yl)-N-[(3-chlorophenyl)methyl]-N-[4-(dimethylamino)phenyl]acetamide | 2-(1H-benzotriazol-1-yl)-N-[(3-chlorophenyl)methyl]-N-[4-(dimethylamino)phenyl]acetamide | XDS | 99.80% | 3.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 13.4 | - | - | - | |||||
| 7ggg | 1.74 Å | 2023-11-08 | X-ray | OVX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.86% | 22.81% | 6.80% | 1-(3-chlorophenyl)-4-(isoquinoline-4-carbonyl)piperazin-2-one | 1-(3-chlorophenyl)-4-(isoquinoline-4-carbonyl)piperazin-2-one | XDS | 99.70% | 16.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 56.9 | - | - | - | |||||
| 7ggh | 1.68 Å | 2023-11-08 | X-ray | OWC | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.15% | 24.64% | 9.70% | (3S)-3-(3-chlorophenyl)-1-(isoquinolin-4-yl)piperidin-2-one | (3S)-3-(3-chlorophenyl)-1-(isoquinolin-4-yl)piperidin-2-one | XDS | 97.70% | 10.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 38.7 | - | - | - | |||||
| 7ggi | 1.65 Å | 2023-11-08 | X-ray | OWX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.27% | 20.97% | 7.00% | 2-(3-chlorophenyl)-N-(1,2,3,4-tetrahydro-1,7-naphthyridin-5-yl)acetamide | 2-(3-chlorophenyl)-N-(1,2,3,4-tetrahydro-1,7-naphthyridin-5-yl)acetamide | XDS | 99.10% | 15.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 74.4 | - | - | - | |||||
| 7ggj | 1.80 Å | 2023-11-08 | X-ray | OYF | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.28% | 23.39% | 14.70% | (4R)-6-chloro-N-(2,7-naphthyridin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-(2,7-naphthyridin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 99.70% | 4.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 51.3 | - | - | - | |||||
| 7ggk | 1.73 Å | 2023-11-08 | X-ray | OYX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.00% | 22.53% | 11.50% | N-(2-cyclohexylethyl)-2-(isoquinolin-4-yl)-N-[(thiophen-2-yl)methyl]acetamide | N-(2-cyclohexylethyl)-2-(isoquinolin-4-yl)-N-[(thiophen-2-yl)methyl]acetamide | XDS | 99.80% | 5.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 59.9 | - | - | - | |||||
| 7ggl | 2.22 Å | 2023-11-08 | X-ray | OZC | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.20% | 27.23% | 10.00% | (3S)-3-(4-chlorophenyl)-1-(isoquinolin-4-yl)piperidin-2-one | (3S)-3-(4-chlorophenyl)-1-(isoquinolin-4-yl)piperidin-2-one | XDS | 99.60% | 11.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 18.2 | - | - | - | |||||
| 7ggm | 1.84 Å | 2023-11-08 | X-ray | OZX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.21% | 23.51% | 9.00% | 2-(6-chloropyridin-2-yl)-N-(isoquinolin-4-yl)acetamide | 2-(6-chloropyridin-2-yl)-N-(isoquinolin-4-yl)acetamide | XDS | 99.60% | 12.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 49.8 | - | - | - | |||||
| 7ggn | 1.93 Å | 2023-11-08 | X-ray | P0X | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.49% | 23.89% | 8.00% | 4-[4-(3-chlorophenyl)-3-oxopiperazine-1-carbonyl]quinolin-2(1H)-one | 4-[4-(3-chlorophenyl)-3-oxopiperazine-1-carbonyl]quinolin-2(1H)-one | XDS | 99.70% | 14.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 46.1 | - | - | - | |||||
| 7ggo | 1.69 Å | 2023-11-08 | X-ray | P3L | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.58% | 22.94% | 6.90% | (4R)-6-chloro-N-[(4R)-2-oxopiperidin-4-yl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-[(4R)-2-oxopiperidin-4-yl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 99.10% | 14.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 55.6 | - | - | - | |||||
| 7ggp | 1.90 Å | 2023-11-08 | X-ray | P4R | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.62% | 21.79% | 10.00% | (1'M,4S)-6-chloro-1'-(isoquinolin-4-yl)-2,3-dihydrospiro[[1]benzopyran-4,4'-imidazolidine]-2',5'-dione | (1'M,4S)-6-chloro-1'-(isoquinolin-4-yl)-2,3-dihydrospiro[[1]benzopyran-4,4'-imidazolidine]-2',5'-dione | XDS | 99.80% | 9.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 67.2 | - | - | - | |||||
| 7ggq | 1.81 Å | 2023-11-08 | X-ray | OQL | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.97% | 22.70% | 8.20% | (4R)-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide | (4R)-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide | XDS | 99.70% | 9.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 58.0 | - | - | - | |||||
| 7ggr | 1.87 Å | 2023-11-08 | X-ray | P6O | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.47% | 22.75% | 10.50% | (4S)-6-chloro-4-hydroxy-N-(isoquinolin-4-yl)-2-oxo-1,2,3,4-tetrahydroquinoline-4-carboxamide | (4S)-6-chloro-4-hydroxy-N-(isoquinolin-4-yl)-2-oxo-1,2,3,4-tetrahydroquinoline-4-carboxamide | XDS | 99.20% | 7.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 57.6 | - | - | - | |||||
| 7ggs | 1.49 Å | 2023-11-08 | X-ray | P7R | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.99% | 22.02% | 10.10% | (4R)-6-chloro-N-(1-methyl-2-oxo-2,3-dihydro-1H-imidazo[4,5-c]pyridin-7-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-(1-methyl-2-oxo-2,3-dihydro-1H-imidazo[4,5-c]pyridin-7-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 99.60% | 6.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 64.8 | - | - | - | |||||
| 7ggt | 1.82 Å | 2023-11-08 | X-ray | P9O | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.55% | 22.53% | 11.60% | (4R)-6-chloro-N-(5,6,7,8-tetrahydro-2,6-naphthyridin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-(5,6,7,8-tetrahydro-2,6-naphthyridin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 99.70% | 8.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 59.9 | - | - | - | |||||
| 7ggu | 2.29 Å | 2023-11-08 | X-ray | PJ6 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.65% | 23.72% | 14.10% | (4S)-6-chloro-N-(isoquinolin-4-yl)-4-methoxy-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-4-methoxy-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 99.60% | 6.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 47.6 | - | - | - | |||||
| 7ggv | 1.97 Å | 2023-11-08 | X-ray | PJX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.04% | 23.50% | 19.30% | (4R)-6-chloro-N-(isoquinolin-4-yl)-1-methyl-1,2,3,4-tetrahydroquinoline-4-carboxamide | (4R)-6-chloro-N-(isoquinolin-4-yl)-1-methyl-1,2,3,4-tetrahydroquinoline-4-carboxamide | XDS | 99.80% | 5.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 49.8 | - | - | - | |||||
| 7ggw | 1.92 Å | 2023-11-08 | X-ray | PKW | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.21% | 24.01% | 25.70% | (4R)-6-chloro-N-(1-methyl-1H-imidazo[4,5-c]pyridin-7-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-(1-methyl-1H-imidazo[4,5-c]pyridin-7-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 99.90% | 3.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 44.7 | - | - | - | |||||
| 7ggx | 1.85 Å | 2023-11-08 | X-ray | PQ6 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.82% | 22.77% | 12.50% | (4R)-4-(aminomethyl)-6-chloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-4-(aminomethyl)-6-chloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 99.90% | 6.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 57.5 | - | - | - | |||||
| 7ggy | 2.17 Å | 2023-11-08 | X-ray | PUU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.30% | 27.81% | 20.90% | (4R)-1-acetyl-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide | (4R)-1-acetyl-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide | XDS | 99.70% | 4.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 15.0 | - | - | - | |||||
| 7ggz | 1.46 Å | 2023-11-08 | X-ray | PVR | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.67% | 20.91% | 9.50% | 2-(4-acetylpiperazin-1-yl)-N-(4-cyclopropylpyridin-3-yl)acetamide | 2-(4-acetylpiperazin-1-yl)-N-(4-cyclopropylpyridin-3-yl)acetamide | XDS | 97.50% | 7.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 74.7 | - | - | - | |||||
| 7gh0 | 1.69 Å | 2023-11-08 | X-ray | PWR | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.69% | 21.95% | 10.70% | (4~{R})-6-chloranyl-~{N}-(2-oxidanylisoquinolin-4-yl)-3,4-dihydro-2~{H}-chromene-4-carboxamide | (4~{R})-6-chloranyl-~{N}-(2-oxidanylisoquinolin-4-yl)-3,4-dihydro-2~{H}-chromene-4-carboxamide | XDS | 99.70% | 7.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 65.6 | - | - | - | |||||
| 7gh1 | 1.52 Å | 2023-11-08 | X-ray | PZ6 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.34% | 21.38% | 8.40% | (4S)-4-(aminomethyl)-6-chloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4S)-4-(aminomethyl)-6-chloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 99.80% | 7.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 70.9 | - | - | - | |||||
| 7gh2 | 1.62 Å | 2023-11-08 | X-ray | Q0I | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.60% | 21.81% | 11.80% | (4R)-6-chloro-N-(isoquinolin-4-yl)-N-propanoyl-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-(isoquinolin-4-yl)-N-propanoyl-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 98.80% | 6.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 66.9 | - | - | - | |||||
| 7gh3 | 1.90 Å | 2023-11-08 | X-ray | Q1C | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.03% | 23.68% | 22.30% | 2-(4-methylpiperidin-1-yl)-N-(4-methylpyridin-3-yl)acetamide | 2-(4-methylpiperidin-1-yl)-N-(4-methylpyridin-3-yl)acetamide | XDS | 99.60% | 4.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 48.2 | - | - | - | |||||
| 7gh4 | 2.27 Å | 2023-11-08 | X-ray | Q1U | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.29% | 24.83% | 15.30% | 2-(3-chlorophenyl)-N-(6-methoxyisoquinolin-4-yl)acetamide | 2-(3-chlorophenyl)-N-(6-methoxyisoquinolin-4-yl)acetamide | XDS | 99.30% | 6.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 36.8 | - | - | - | |||||
| 7gh5 | 1.66 Å | 2023-11-08 | X-ray | Q2G | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.96% | 21.27% | 11.80% | N-(3-chlorophenyl)-N'-(4-methylpyridin-3-yl)urea | N-(3-chlorophenyl)-N'-(4-methylpyridin-3-yl)urea | XDS | 99.30% | 5.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 71.8 | - | - | - | |||||
| 7gh6 | 1.57 Å | 2023-11-08 | X-ray | Q2U | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.13% | 21.56% | 8.90% | N-[2-(2-methoxyphenoxy)ethyl]-2-oxo-1,2-dihydroquinoline-4-carboxamide | N-[2-(2-methoxyphenoxy)ethyl]-2-oxo-1,2-dihydroquinoline-4-carboxamide | XDS | 96.20% | 9.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 69.3 | - | - | - | |||||
| 7gh7 | 1.67 Å | 2023-11-08 | X-ray | Q36 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.01% | 20.43% | 12.60% | (2R)-2-(3-cyanophenyl)-N-(4-methylpyridin-3-yl)propanamide | (2R)-2-(3-cyanophenyl)-N-(4-methylpyridin-3-yl)propanamide | XDS | 99.70% | 6.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 78.6 | - | - | - | |||||
| 7gh8 | 1.52 Å | 2023-11-08 | X-ray | Q3U | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 15.85% | 20.75% | 8.40% | N-(5-aminopyridin-3-yl)-N'-(3-chlorophenyl)urea | N-(5-aminopyridin-3-yl)-N'-(3-chlorophenyl)urea | XDS | 97.80% | 6.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 76.2 | - | - | - | |||||
| 7gh9 | 1.57 Å | 2023-11-08 | X-ray | Q45 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.93% | 21.06% | 9.90% | 2-[(2S)-2-{2-[(methanesulfonyl)amino]ethyl}piperidin-1-yl]-N-(pyridin-3-yl)acetamide | 2-[(2S)-2-{2-[(methanesulfonyl)amino]ethyl}piperidin-1-yl]-N-(pyridin-3-yl)acetamide | XDS | 99.20% | 7.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 73.6 | - | - | - | |||||
| 7gha | 1.72 Å | 2023-11-08 | X-ray | Q4F | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.21% | 22.52% | 8.90% | 1-{(1S,4S)-5-[(3-chlorophenyl)methyl]-2,5-diazabicyclo[2.2.1]heptan-2-yl}ethan-1-one | 1-{(1S,4S)-5-[(3-chlorophenyl)methyl]-2,5-diazabicyclo[2.2.1]heptan-2-yl}ethan-1-one | XDS | 99.50% | 9.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 60.0 | - | - | - | |||||
| 7ghb | 1.56 Å | 2023-11-08 | X-ray | Q4R | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.12% | 22.47% | 9.20% | N~2~-methyl-N-(4-methylpyridin-3-yl)-N~2~-(quinoline-8-sulfonyl)glycinamide | N~2~-methyl-N-(4-methylpyridin-3-yl)-N~2~-(quinoline-8-sulfonyl)glycinamide | XDS | 98.20% | 7.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 60.6 | - | - | - | |||||
| 7ghc | 1.48 Å | 2023-11-08 | X-ray | Q5C | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.87% | 20.97% | 8.40% | N-(4-methylpyridin-3-yl)-N~2~-[(pyridin-3-yl)acetyl]glycinamide | N-(4-methylpyridin-3-yl)-N~2~-[(pyridin-3-yl)acetyl]glycinamide | XDS | 98.10% | 7.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 74.4 | - | - | - | |||||
| 7ghd | 1.97 Å | 2023-11-08 | X-ray | Q5K | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.09% | 23.44% | 7.60% | (2S)-N-{2-[(4-fluorobenzene-1-sulfonyl)amino]phenyl}-2-hydroxy-2-(pyridin-3-yl)acetamide | (2S)-N-{2-[(4-fluorobenzene-1-sulfonyl)amino]phenyl}-2-hydroxy-2-(pyridin-3-yl)acetamide | XDS | 99.60% | 9.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 50.6 | - | - | - | |||||
| 7ghe | 2.17 Å | 2023-11-08 | X-ray | Q5R | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.28% | 25.63% | 11.40% | 4-(4-phenylpiperazine-1-carbonyl)quinolin-2(1H)-one | 4-(4-phenylpiperazine-1-carbonyl)quinolin-2(1H)-one | XDS | 99.80% | 8.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 29.6 | - | - | - | |||||
| 7ghf | 1.66 Å | 2023-11-08 | X-ray | Q60 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.23% | 22.31% | 8.80% | N'-[(1-methyl-1H-1,2,3-triazol-4-yl)methyl]-N-(2-phenylethyl)-N-[(pyridin-3-yl)methyl]urea | N'-[(1-methyl-1H-1,2,3-triazol-4-yl)methyl]-N-(2-phenylethyl)-N-[(pyridin-3-yl)methyl]urea | XDS | 99.40% | 8.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 62.0 | - | - | - | |||||
| 7ghg | 1.45 Å | 2023-11-08 | X-ray | Q69 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.90% | 20.90% | 8.90% | N-[(1R)-1-(3-chlorophenyl)-2-hydroxyethyl]acetamide | N-[(1R)-1-(3-chlorophenyl)-2-hydroxyethyl]acetamide | XDS | 98.80% | 7.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 74.8 | - | - | - | |||||
| 7ghh | 1.56 Å | 2023-11-08 | X-ray | Q6U | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.18% | 22.34% | 8.50% | 1-[4-(prop-2-yn-1-yl)piperazin-1-yl]ethan-1-one | 1-[4-(prop-2-yn-1-yl)piperazin-1-yl]ethan-1-one | XDS | 98.90% | 8.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 61.8 | - | - | - | |||||
| 7ghi | 1.65 Å | 2023-11-08 | X-ray | Q7C | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 18.08% | 21.88% | 9.60% | N~3~-acetyl-N~3~-[(3S)-1,1-dioxo-1lambda~6~-thiolan-3-yl]-beta-alaninamide | N~3~-acetyl-N~3~-[(3S)-1,1-dioxo-1lambda~6~-thiolan-3-yl]-beta-alaninamide | XDS | 99.70% | 8.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 66.3 | - | - | - | |||||
| 7ghj | 1.53 Å | 2023-11-08 | X-ray | Q7R | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 15.60% | 20.93% | 6.10% | 1-[4-(diphenylmethyl)piperazin-1-yl]ethan-1-one | 1-[4-(diphenylmethyl)piperazin-1-yl]ethan-1-one | XDS | 93.30% | 10.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 74.6 | - | - | - | |||||
| 7ghk | 1.52 Å | 2023-11-08 | X-ray | Q8I | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.51% | 20.79% | 8.80% | 1-(5-fluoro-1H-indol-3-yl)-N-methylmethanamine | 1-(5-fluoro-1H-indol-3-yl)-N-methylmethanamine | XDS | 99.10% | 8.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 75.9 | - | - | - | |||||
| 7ghl | 1.41 Å | 2023-11-08 | X-ray | Q8O | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 17.41% | 20.46% | 7.10% | 2-(1H-benzimidazol-6-yl)-N-(4-methylpyridin-3-yl)acetamide | 2-(1H-benzimidazol-6-yl)-N-(4-methylpyridin-3-yl)acetamide | XDS | 98.10% | 9.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 78.5 | - | - | - | |||||
| 7ghm | 1.69 Å | 2023-11-08 | X-ray | Q99 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.54% | 23.43% | 9.80% | N-[2-(3-chloro-5-{[(2S)-4-oxoazetidin-2-yl]oxy}phenoxy)ethyl]-2-oxo-1,2-dihydroquinoline-4-carboxamide | N-[2-(3-chloro-5-{[(2S)-4-oxoazetidin-2-yl]oxy}phenoxy)ethyl]-2-oxo-1,2-dihydroquinoline-4-carboxamide | XDS | 56.90% | 11.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 50.7 | - | - | - | |||||
| 7ghn | 1.73 Å | 2023-11-08 | X-ray | QBR | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.57% | 25.15% | 14.70% | N-(4-tert-butylphenyl)-N-[(1R)-2-{[2-(3-fluorophenyl)ethyl]amino}-2-oxo-1-(pyridin-3-yl)ethyl]furan-2-carboxamide | N-(4-tert-butylphenyl)-N-[(1R)-2-{[2-(3-fluorophenyl)ethyl]amino}-2-oxo-1-(pyridin-3-yl)ethyl]furan-2-carboxamide | XDS | 68.60% | 7.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 33.9 | - | - | - | |||||
| 7gho | 1.86 Å | 2023-11-08 | X-ray | QC3 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.99% | 27.90% | 19.80% | (3S)-3-(3-chlorophenyl)-1-(isoquinolin-4-yl)pyrrolidin-2-one | (3S)-3-(3-chlorophenyl)-1-(isoquinolin-4-yl)pyrrolidin-2-one | XDS | 59.90% | 5.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 14.5 | - | - | - | |||||
| 7ghp | 1.54 Å | 2023-11-08 | X-ray | OYF | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.16% | 24.71% | 8.80% | (4R)-6-chloro-N-(2,7-naphthyridin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-(2,7-naphthyridin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 62.70% | 11.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 38.0 | - | - | - | |||||
| 7ghq | 1.72 Å | 2023-11-08 | X-ray | OYX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.90% | 25.09% | 10.40% | N-(2-cyclohexylethyl)-2-(isoquinolin-4-yl)-N-[(thiophen-2-yl)methyl]acetamide | N-(2-cyclohexylethyl)-2-(isoquinolin-4-yl)-N-[(thiophen-2-yl)methyl]acetamide | XDS | 62.60% | 11.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 34.5 | - | - | - | |||||
| 7ghr | 1.66 Å | 2023-11-08 | X-ray | OZC | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.69% | 26.03% | 9.70% | (3S)-3-(4-chlorophenyl)-1-(isoquinolin-4-yl)piperidin-2-one | (3S)-3-(4-chlorophenyl)-1-(isoquinolin-4-yl)piperidin-2-one | XDS | 57.40% | 11.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 26.3 | - | - | - | |||||
| 7ghs | 1.66 Å | 2023-11-08 | X-ray | OVX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.66% | 24.84% | 11.20% | 1-(3-chlorophenyl)-4-(isoquinoline-4-carbonyl)piperazin-2-one | 1-(3-chlorophenyl)-4-(isoquinoline-4-carbonyl)piperazin-2-one | XDS | 59.90% | 10.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 36.7 | - | - | - | |||||
| 7ght | 1.79 Å | 2023-11-08 | X-ray | OWC | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.46% | 25.67% | 12.10% | (3S)-3-(3-chlorophenyl)-1-(isoquinolin-4-yl)piperidin-2-one | (3S)-3-(3-chlorophenyl)-1-(isoquinolin-4-yl)piperidin-2-one | XDS | 73.20% | 9.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 29.4 | - | - | - | |||||
| 7ghu | 1.64 Å | 2023-11-08 | X-ray | P4R | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.21% | 25.57% | 12.10% | (1'M,4S)-6-chloro-1'-(isoquinolin-4-yl)-2,3-dihydrospiro[[1]benzopyran-4,4'-imidazolidine]-2',5'-dione | (1'M,4S)-6-chloro-1'-(isoquinolin-4-yl)-2,3-dihydrospiro[[1]benzopyran-4,4'-imidazolidine]-2',5'-dione | XDS | 63.60% | 10.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 30.3 | - | - | - | |||||
| 7ghv | 1.63 Å | 2023-11-08 | X-ray | OZX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.27% | 25.52% | 10.40% | 2-(6-chloropyridin-2-yl)-N-(isoquinolin-4-yl)acetamide | 2-(6-chloropyridin-2-yl)-N-(isoquinolin-4-yl)acetamide | XDS | 66.80% | 10.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 30.5 | - | - | - | |||||
| 7ghw | 1.58 Å | 2023-11-08 | X-ray | P3L | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.70% | 24.57% | 8.80% | (4R)-6-chloro-N-[(4R)-2-oxopiperidin-4-yl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-[(4R)-2-oxopiperidin-4-yl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 69.40% | 11.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 39.5 | - | - | - | |||||
| 7ghx | 1.49 Å | 2023-11-08 | X-ray | P7R | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.36% | 24.15% | 8.40% | (4R)-6-chloro-N-(1-methyl-2-oxo-2,3-dihydro-1H-imidazo[4,5-c]pyridin-7-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-(1-methyl-2-oxo-2,3-dihydro-1H-imidazo[4,5-c]pyridin-7-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 67.00% | 11.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 43.5 | - | - | - | |||||
| 7ghy | 1.62 Å | 2023-11-08 | X-ray | P9O | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.57% | 24.40% | 9.50% | (4R)-6-chloro-N-(5,6,7,8-tetrahydro-2,6-naphthyridin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-(5,6,7,8-tetrahydro-2,6-naphthyridin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 63.60% | 11.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 41.0 | - | - | - | |||||
| 7ghz | 1.67 Å | 2023-11-08 | X-ray | PJX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.35% | 25.41% | 8.70% | (4R)-6-chloro-N-(isoquinolin-4-yl)-1-methyl-1,2,3,4-tetrahydroquinoline-4-carboxamide | (4R)-6-chloro-N-(isoquinolin-4-yl)-1-methyl-1,2,3,4-tetrahydroquinoline-4-carboxamide | XDS | 60.50% | 12.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 31.5 | - | - | - | |||||
| 7gi0 | 1.58 Å | 2023-11-08 | X-ray | OQL | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.85% | 25.07% | 9.30% | (4R)-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide | (4R)-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide | XDS | 65.10% | 11.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 34.6 | - | - | - | |||||
| 7gi1 | 1.62 Å | 2023-11-08 | X-ray | PJ6 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.42% | 25.74% | 9.70% | (4S)-6-chloro-N-(isoquinolin-4-yl)-4-methoxy-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-4-methoxy-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 63.30% | 11.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 28.8 | - | - | - | |||||
| 7gi2 | 1.61 Å | 2023-11-08 | X-ray | PQ6 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.53% | 24.30% | 8.40% | (4R)-4-(aminomethyl)-6-chloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-4-(aminomethyl)-6-chloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 68.80% | 12.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 41.9 | - | - | - | |||||
| 7gi3 | 2.10 Å | 2023-11-08 | X-ray | Q0I, CA | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.07% | 26.22% | 16.10% | (4R)-6-chloro-N-(isoquinolin-4-yl)-N-propanoyl-3,4-dihydro-2H-1-benzopyran-4-carboxamide; CALCIUM ION | (4R)-6-chloro-N-(isoquinolin-4-yl)-N-propanoyl-3,4-dihydro-2H-1-benzopyran-4-carboxamide; CALCIUM ION | XDS | 56.00% | 8.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | CA | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 25.0 | - | - | - | |||||
| 7gi4 | 1.72 Å | 2023-11-08 | X-ray | QCC | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.17% | 25.40% | 12.10% | 2-(3-chloro-5-{[(2S)-4-oxoazetidin-2-yl]oxy}phenyl)-N-(4-methylpyridin-3-yl)acetamide | 2-(3-chloro-5-{[(2S)-4-oxoazetidin-2-yl]oxy}phenyl)-N-(4-methylpyridin-3-yl)acetamide | XDS | 65.70% | 11.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 31.6 | - | - | - | |||||
| 7gi5 | 2.00 Å | 2023-11-08 | X-ray | QCO | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 24.89% | 29.35% | 17.30% | N-[(1R)-2-{[2-(3-fluorophenyl)ethyl]amino}-2-oxo-1-(pyridin-3-yl)ethyl]-N-{4-[(propan-2-yl)oxy]phenyl}-1H-imidazole-4-carboxamide | N-[(1R)-2-{[2-(3-fluorophenyl)ethyl]amino}-2-oxo-1-(pyridin-3-yl)ethyl]-N-{4-[(propan-2-yl)oxy]phenyl}-1H-imidazole-4-carboxamide | XDS | 54.90% | 8.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 9.0 | - | - | - | |||||
| 7gi6 | 1.69 Å | 2023-11-08 | X-ray | QD4 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.35% | 26.45% | 13.40% | (4R)-6-chloro-N~4~-(isoquinolin-4-yl)-3,4-dihydroquinoline-1,4(2H)-dicarboxamide | (4R)-6-chloro-N~4~-(isoquinolin-4-yl)-3,4-dihydroquinoline-1,4(2H)-dicarboxamide | XDS | 63.80% | 8.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 23.3 | - | - | - | |||||
| 7gi7 | 1.74 Å | 2023-11-08 | X-ray | QD9 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.06% | 25.64% | 12.30% | N-[(3-chlorophenyl)methyl]-N-[5-(dimethylamino)pyridin-2-yl]-2-(isoquinolin-4-yl)acetamide | N-[(3-chlorophenyl)methyl]-N-[5-(dimethylamino)pyridin-2-yl]-2-(isoquinolin-4-yl)acetamide | XDS | 60.80% | 8.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 29.6 | - | - | - | |||||
| 7gi8 | 1.80 Å | 2023-11-08 | X-ray | QDF | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.41% | 25.56% | 12.30% | (4R)-6-chloro-N-(isoquinolin-4-yl)-2-oxo-1,2,3,4-tetrahydroquinoline-4-carboxamide | (4R)-6-chloro-N-(isoquinolin-4-yl)-2-oxo-1,2,3,4-tetrahydroquinoline-4-carboxamide | XDS | 64.80% | 9.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 30.3 | - | - | - | |||||
| 7gi9 | 1.91 Å | 2023-11-08 | X-ray | QDU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.94% | 28.20% | 13.50% | 2-(2,5-difluorophenyl)-N-(isoquinolin-4-yl)acetamide | 2-(2,5-difluorophenyl)-N-(isoquinolin-4-yl)acetamide | XDS | 54.90% | 10.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 13.1 | - | - | - | |||||
| 7gia | 2.04 Å | 2023-11-08 | X-ray | QE3 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.11% | 26.86% | 16.30% | 2-(5-chloropyridin-2-yl)-N-(isoquinolin-4-yl)acetamide | 2-(5-chloropyridin-2-yl)-N-(isoquinolin-4-yl)acetamide | XDS | 56.40% | 8.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 20.5 | - | - | - | |||||
| 7gib | 1.68 Å | 2023-11-08 | X-ray | QER | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.87% | 25.65% | 10.10% | 2-(3-fluorophenyl)-N-(isoquinolin-4-yl)acetamide | 2-(3-fluorophenyl)-N-(isoquinolin-4-yl)acetamide | XDS | 57.30% | 11.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 29.5 | - | - | - | |||||
| 7gic | 1.92 Å | 2023-11-08 | X-ray | QEX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 29.34% | 32.87% | 23.50% | N-(isoquinolin-4-yl)-2-(3-methylphenyl)acetamide | N-(isoquinolin-4-yl)-2-(3-methylphenyl)acetamide | XDS | 78.20% | 7.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 4.5 | - | - | - | |||||
| 7gid | 1.90 Å | 2023-11-08 | X-ray | QF5 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.67% | 26.58% | 15.00% | N-(isoquinolin-4-yl)-2-phenylacetamide | N-(isoquinolin-4-yl)-2-phenylacetamide | XDS | 61.50% | 9.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 22.5 | - | - | - | |||||
| 7gie | 2.17 Å | 2023-11-08 | X-ray | QF9 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 24.19% | 29.23% | 23.30% | 2-(3-chloro-5-cyanophenyl)-N-(isoquinolin-4-yl)acetamide | 2-(3-chloro-5-cyanophenyl)-N-(isoquinolin-4-yl)acetamide | XDS | 50.70% | 6.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 9.2 | - | - | - | |||||
| 7gif | 1.68 Å | 2023-11-08 | X-ray | QFL | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.47% | 25.58% | 9.20% | 2-(4-fluorophenyl)-N-(isoquinolin-4-yl)acetamide | 2-(4-fluorophenyl)-N-(isoquinolin-4-yl)acetamide | XDS | 58.80% | 11.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 30.2 | - | - | - | |||||
| 7gig | 1.94 Å | 2023-11-08 | X-ray | QFU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.80% | 28.00% | 18.10% | 2-(3-cyanophenyl)-N-(isoquinolin-4-yl)acetamide | 2-(3-cyanophenyl)-N-(isoquinolin-4-yl)acetamide | XDS | 58.60% | 8.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 14.0 | - | - | - | |||||
| 7gih | 1.63 Å | 2023-11-08 | X-ray | QG3 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.90% | 25.27% | 9.70% | 2-(3,5-difluorophenyl)-N-(isoquinolin-4-yl)acetamide | 2-(3,5-difluorophenyl)-N-(isoquinolin-4-yl)acetamide | XDS | 69.20% | 10.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 32.9 | - | - | - | |||||
| 7gii | 2.12 Å | 2023-11-08 | X-ray | QGC | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.65% | 28.19% | 21.60% | (4R)-6-chloro-N-(isoquinolin-4-yl)-1-[(4H-1,2,4-triazol-3-yl)methyl]-1,2,3,4-tetrahydroquinoline-4-carboxamide | (4R)-6-chloro-N-(isoquinolin-4-yl)-1-[(4H-1,2,4-triazol-3-yl)methyl]-1,2,3,4-tetrahydroquinoline-4-carboxamide | XDS | 57.90% | 7.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 13.3 | - | - | - | |||||
| 7gij | 1.90 Å | 2023-11-08 | X-ray | QGO | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.94% | 28.05% | 16.50% | (4S)-6-chloro-4-[2-(dimethylamino)-2-oxoethyl]-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4S)-6-chloro-4-[2-(dimethylamino)-2-oxoethyl]-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 59.50% | 7.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 13.8 | - | - | - | |||||
| 7gik | 2.09 Å | 2023-11-08 | X-ray | QGX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.37% | 28.59% | 16.10% | (2R)-2-amino-2-(3,4-dichlorophenyl)-N-(isoquinolin-4-yl)acetamide | (2R)-2-amino-2-(3,4-dichlorophenyl)-N-(isoquinolin-4-yl)acetamide | XDS; XDS; XDS | 61.30% | 8.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 11.6 | - | - | - | |||||
| 7gil | 2.07 Å | 2023-11-08 | X-ray | QH6 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.56% | 28.73% | 16.40% | 2-(3-chloro-4-fluorophenyl)-N-(isoquinolin-4-yl)acetamide | 2-(3-chloro-4-fluorophenyl)-N-(isoquinolin-4-yl)acetamide | XDS | 56.50% | 8.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 10.9 | - | - | - | |||||
| 7gim | 2.03 Å | 2023-11-08 | X-ray | QHI | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.62% | 27.86% | 15.30% | (4R)-6,8-dichloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6,8-dichloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 52.50% | 8.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 14.8 | - | - | - | |||||
| 7gin | 1.86 Å | 2023-11-08 | X-ray | QHU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.22% | 25.39% | 14.50% | (4R)-6,7-dichloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6,7-dichloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 59.00% | 9.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 31.9 | - | - | - | |||||
| 7gio | 2.04 Å | 2023-11-08 | X-ray | QI4 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.69% | 26.17% | 17.20% | (2S)-2-(3,4-dichlorophenyl)-N-(isoquinolin-4-yl)propanamide | (2S)-2-(3,4-dichlorophenyl)-N-(isoquinolin-4-yl)propanamide | XDS | 75.70% | 7.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 25.4 | - | - | - | |||||
| 7gip | 2.05 Å | 2023-11-08 | X-ray | QI7 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.22% | 26.20% | 16.10% | (4R)-6-chloro-1-[(1H-imidazol-2-yl)methyl]-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide | (4R)-6-chloro-1-[(1H-imidazol-2-yl)methyl]-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide | XDS | 61.90% | 9.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 25.0 | - | - | - | |||||
| 7giq | 1.92 Å | 2023-11-08 | X-ray | QIM | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.11% | 25.97% | 15.20% | (4R)-6-chloro-N-[6-(methanesulfonyl)isoquinolin-4-yl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-[6-(methanesulfonyl)isoquinolin-4-yl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 77.40% | 8.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 26.9 | - | - | - | |||||
| 7gir | 2.18 Å | 2023-11-08 | X-ray | QIB | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.23% | 26.68% | 18.00% | (4S)-6-chloro-4-hydroxy-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4S)-6-chloro-4-hydroxy-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 61.70% | 8.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 21.8 | - | - | - | |||||
| 7gis | 1.84 Å | 2023-11-08 | X-ray | QIQ | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.36% | 25.99% | 15.30% | 2-(5-chloropyridin-3-yl)-N-(isoquinolin-4-yl)acetamide | 2-(5-chloropyridin-3-yl)-N-(isoquinolin-4-yl)acetamide | XDS | 72.30% | 8.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 26.8 | - | - | - | |||||
| 7git | 2.06 Å | 2023-11-08 | X-ray | QIT | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.69% | 26.93% | 25.90% | (2S)-2-(3,4-dichlorophenyl)-N-(isoquinolin-4-yl)-2-(methylamino)acetamide | (2S)-2-(3,4-dichlorophenyl)-N-(isoquinolin-4-yl)-2-(methylamino)acetamide | XDS | 66.00% | 6.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 20.0 | - | - | - | |||||
| 7giu | 2.00 Å | 2023-11-08 | X-ray | QIZ | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.34% | 27.01% | 38.10% | (4R)-6-chloro-7-fluoro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-7-fluoro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 66.90% | 4.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 19.4 | - | - | - | |||||
| 7giv | 2.20 Å | 2023-11-08 | X-ray | QJ6 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.79% | 29.92% | 28.20% | (2R)-2-(3,4-dichlorophenyl)-N-(isoquinolin-4-yl)-2-methoxyacetamide | (2R)-2-(3,4-dichlorophenyl)-N-(isoquinolin-4-yl)-2-methoxyacetamide | XDS | 57.10% | 6.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 7.4 | - | - | - | |||||
| 7giw | 2.45 Å | 2023-11-08 | X-ray | QJF | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.02% | 28.31% | 21.10% | (4R)-6-chloro-N-(6-methoxyisoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-(6-methoxyisoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 51.30% | 6.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 12.6 | - | - | - | |||||
| 7gix | 2.07 Å | 2023-11-08 | X-ray | P4R | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.79% | 27.46% | 18.80% | (1'M,4S)-6-chloro-1'-(isoquinolin-4-yl)-2,3-dihydrospiro[[1]benzopyran-4,4'-imidazolidine]-2',5'-dione | (1'M,4S)-6-chloro-1'-(isoquinolin-4-yl)-2,3-dihydrospiro[[1]benzopyran-4,4'-imidazolidine]-2',5'-dione | XDS | 50.60% | 8.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 16.9 | - | - | - | |||||
| 7giy | 2.04 Å | 2023-11-08 | X-ray | OI4 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 25.03% | 29.32% | 19.90% | (4R)-6-chloro-N-(4-methylpyridin-3-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-(4-methylpyridin-3-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 56.80% | 7.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 9.0 | - | - | - | |||||
| 7giz | 1.97 Å | 2023-11-08 | X-ray | PUU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.99% | 27.54% | 17.90% | (4R)-1-acetyl-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide | (4R)-1-acetyl-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide | XDS | 62.80% | 8.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 16.5 | - | - | - | |||||
| 7gj0 | 1.81 Å | 2023-11-08 | X-ray | QJL | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.81% | 26.71% | 10.50% | (4R)-6-chloro-4-{[(N,N-dimethylglycyl)amino]methyl}-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-4-{[(N,N-dimethylglycyl)amino]methyl}-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 58.30% | 11.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 21.4 | - | - | - | |||||
| 7gj1 | 1.95 Å | 2023-11-08 | X-ray | QJR | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.18% | 27.12% | 14.90% | 2-(3,4-dichlorophenyl)-N-(2,7-naphthyridin-4-yl)acetamide | 2-(3,4-dichlorophenyl)-N-(2,7-naphthyridin-4-yl)acetamide | XDS; XDS; XDS | 65.40% | 8.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 18.8 | - | - | - | |||||
| 7gj2 | 1.87 Å | 2023-11-08 | X-ray | QK3 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.74% | 26.38% | 13.20% | (4R)-6-chloro-4-{[2-(1H-imidazol-1-yl)acetamido]methyl}-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-4-{[2-(1H-imidazol-1-yl)acetamido]methyl}-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 73.10% | 9.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 24.0 | - | - | - | |||||
| 7gj3 | 1.92 Å | 2023-11-08 | X-ray | PJ6 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.81% | 25.95% | 12.40% | (4S)-6-chloro-N-(isoquinolin-4-yl)-4-methoxy-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-4-methoxy-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 66.90% | 9.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 27.0 | - | - | - | |||||
| 7gj4 | 2.13 Å | 2023-11-08 | X-ray | QKB | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.79% | 28.26% | 21.40% | (4R)-6-chloro-N-(6-fluoroisoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-(6-fluoroisoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 56.90% | 7.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 12.9 | - | - | - | |||||
| 7gj5 | 2.13 Å | 2023-11-08 | X-ray | PJX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.27% | 27.50% | 22.00% | (4R)-6-chloro-N-(isoquinolin-4-yl)-1-methyl-1,2,3,4-tetrahydroquinoline-4-carboxamide | (4R)-6-chloro-N-(isoquinolin-4-yl)-1-methyl-1,2,3,4-tetrahydroquinoline-4-carboxamide | XDS | 67.60% | 7.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 16.6 | - | - | - | |||||
| 7gj6 | 2.03 Å | 2023-11-08 | X-ray | QKI | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.95% | 26.65% | 16.10% | (2S)-2-(3,4-dichlorophenyl)-2-hydroxy-N-(isoquinolin-4-yl)acetamide | (2S)-2-(3,4-dichlorophenyl)-2-hydroxy-N-(isoquinolin-4-yl)acetamide | XDS | 63.10% | 8.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 22.0 | - | - | - | |||||
| 7gj7 | 1.88 Å | 2023-11-08 | X-ray | Q0I | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.04% | 24.91% | 10.20% | (4R)-6-chloro-N-(isoquinolin-4-yl)-N-propanoyl-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-(isoquinolin-4-yl)-N-propanoyl-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 67.30% | 10.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 36.0 | - | - | - | |||||
| 7gj8 | 2.00 Å | 2023-11-08 | X-ray | QKR | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.08% | 26.72% | 15.00% | 2-(3,4-dichlorophenyl)-2,2-difluoro-N-(isoquinolin-4-yl)acetamide | 2-(3,4-dichlorophenyl)-2,2-difluoro-N-(isoquinolin-4-yl)acetamide | XDS | 75.50% | 8.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 21.4 | - | - | - | |||||
| 7gj9 | 2.09 Å | 2023-11-08 | X-ray | QL3 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 24.47% | 29.93% | 23.30% | N-(6-acetamidopyridin-3-yl)-N-[(3-chlorophenyl)methyl]-2-(isoquinolin-4-yl)acetamide | N-(6-acetamidopyridin-3-yl)-N-[(3-chlorophenyl)methyl]-2-(isoquinolin-4-yl)acetamide | XDS | 47.50% | 7.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 7.4 | - | - | - | |||||
| 7gja | 2.10 Å | 2023-11-08 | X-ray | QLC | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.53% | 28.86% | 17.80% | (2R)-2-(3,4-dichlorophenyl)-N-(isoquinolin-4-yl)-2-(2-methoxyethoxy)acetamide | (2R)-2-(3,4-dichlorophenyl)-N-(isoquinolin-4-yl)-2-(2-methoxyethoxy)acetamide | XDS | 50.60% | 7.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 10.5 | - | - | - | |||||
| 7gjb | 2.03 Å | 2023-11-08 | X-ray | QLO | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.33% | 27.35% | 24.50% | (4S)-6-chloro-N-(isoquinolin-4-yl)-4-[2-(methylamino)-2-oxoethyl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-4-[2-(methylamino)-2-oxoethyl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 59.20% | 6.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 17.5 | - | - | - | |||||
| 7gjc | 1.93 Å | 2023-11-08 | X-ray | QM3 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.98% | 28.58% | 23.20% | (1'P,3'S)-6-chloro-1'-(isoquinolin-4-yl)-2,3-dihydrospiro[[1]benzopyran-4,3'-pyrrolidine]-2',5'-dione | (1'P,3'S)-6-chloro-1'-(isoquinolin-4-yl)-2,3-dihydrospiro[[1]benzopyran-4,3'-pyrrolidine]-2',5'-dione | XDS | 100.00% | 8.9 | Diamond (I03) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 11.6 | - | - | - | |||||
| 7gjd | 1.79 Å | 2023-11-08 | X-ray | QM9 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.72% | 26.20% | 15.70% | (4S)-4-amino-6-chloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4S)-4-amino-6-chloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 100.00% | 10.2 | Diamond (I03) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 25.0 | - | - | - | |||||
| 7gje | 2.03 Å | 2023-11-08 | X-ray | QML | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.01% | 26.67% | 25.60% | (3S)-5-chloro-N-(isoquinolin-4-yl)-N-propanoyl-2,3-dihydro-1-benzofuran-3-carboxamide | (3S)-5-chloro-N-(isoquinolin-4-yl)-N-propanoyl-2,3-dihydro-1-benzofuran-3-carboxamide | XDS | 100.00% | 8.5 | Diamond (I03) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 21.9 | - | - | - | |||||
| 7gjf | 1.96 Å | 2023-11-08 | X-ray | QMX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.51% | 27.35% | 15.90% | (4R)-6-chloro-N-(isoquinolin-4-yl)-4-({[(1-methyl-1H-pyrazol-3-yl)methyl]amino}methyl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-(isoquinolin-4-yl)-4-({[(1-methyl-1H-pyrazol-3-yl)methyl]amino}methyl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 61.40% | 8.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 17.5 | - | - | - | |||||
| 7gjg | 1.99 Å | 2023-11-08 | X-ray | QN9 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.69% | 28.03% | 13.30% | (4S)-6-chloro-4-{2-[4-(3-hydroxypropyl)piperazin-1-yl]-2-oxoethyl}-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4S)-6-chloro-4-{2-[4-(3-hydroxypropyl)piperazin-1-yl]-2-oxoethyl}-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 59.60% | 10.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 13.9 | - | - | - | |||||
| 7gjh | 1.92 Å | 2023-11-08 | X-ray | QNU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.08% | 26.86% | 15.90% | (4S)-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 62.00% | 9.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 20.5 | - | - | - | |||||
| 7gji | 1.59 Å | 2023-11-08 | X-ray | QO0 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.99% | 24.86% | 8.20% | (4S)-6,7-dichloro-N-(isoquinolin-4-yl)-4-methoxy-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4S)-6,7-dichloro-N-(isoquinolin-4-yl)-4-methoxy-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 69.00% | 12.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 36.6 | - | - | - | |||||
| 7gjj | 1.75 Å | 2023-11-08 | X-ray | QOO | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.39% | 26.69% | 13.00% | 2-{3-chloro-5-[4-(ethanesulfonyl)piperazin-1-yl]phenyl}-N-(isoquinolin-4-yl)acetamide | 2-{3-chloro-5-[4-(ethanesulfonyl)piperazin-1-yl]phenyl}-N-(isoquinolin-4-yl)acetamide | XDS | 67.60% | 9.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 21.8 | - | - | - | |||||
| 7gjk | 2.10 Å | 2023-11-08 | X-ray | QOU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 24.08% | 29.04% | 16.80% | (4S)-6-chloro-N-(isoquinolin-4-yl)-4-(2-methoxyethyl)-1,2,3,4-tetrahydroquinoline-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-4-(2-methoxyethyl)-1,2,3,4-tetrahydroquinoline-4-carboxamide | XDS | 56.60% | 8.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 9.8 | - | - | - | |||||
| 7gjl | 2.17 Å | 2023-11-08 | X-ray | QOC | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.74% | 28.11% | 18.20% | (3R)-3-(3,4-dichlorophenyl)-1-(isoquinolin-4-yl)piperidin-2-one | (3R)-3-(3,4-dichlorophenyl)-1-(isoquinolin-4-yl)piperidin-2-one | XDS | 52.40% | 7.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 13.5 | - | - | - | |||||
| 7gjm | 1.92 Å | 2023-11-08 | X-ray | QP0 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.45% | 27.56% | 13.20% | (3R,4R)-6-chloro-N-(isoquinolin-4-yl)-3-methyl-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (3R,4R)-6-chloro-N-(isoquinolin-4-yl)-3-methyl-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 59.60% | 9.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 16.4 | - | - | - | |||||
| 7gjn | 2.06 Å | 2023-11-08 | X-ray | QP6 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.40% | 27.94% | 15.00% | (4S)-6,8-difluoro-N-(isoquinolin-4-yl)-4-methoxy-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4S)-6,8-difluoro-N-(isoquinolin-4-yl)-4-methoxy-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 62.70% | 9.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 14.3 | - | - | - | |||||
| 7gjo | 2.20 Å | 2023-11-08 | X-ray | QIZ | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.23% | 28.32% | 16.80% | (4R)-6-chloro-7-fluoro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-7-fluoro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 56.80% | 8.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 12.6 | - | - | - | |||||
| 7gjp | 1.97 Å | 2023-11-08 | X-ray | QPQ | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.45% | 28.13% | 15.00% | (4S)-6-chloro-7-fluoro-N-(isoquinolin-4-yl)-4-methoxy-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4S)-6-chloro-7-fluoro-N-(isoquinolin-4-yl)-4-methoxy-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 66.00% | 7.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 13.5 | - | - | - | |||||
| 7gjq | 1.97 Å | 2023-11-08 | X-ray | QQ6 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.36% | 27.10% | 11.90% | (4R)-6-chloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzothiopyran-4-carboxamide | (4R)-6-chloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzothiopyran-4-carboxamide | XDS | 65.60% | 11.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 18.9 | - | - | - | |||||
| 7gjr | 2.25 Å | 2023-11-08 | X-ray | QQF | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.91% | 30.27% | 17.90% | 2-(3,4-dichlorophenyl)-N-(isoquinolin-4-yl)-N~2~-(methoxyacetyl)-L-alaninamide | 2-(3,4-dichlorophenyl)-N-(isoquinolin-4-yl)-N~2~-(methoxyacetyl)-L-alaninamide | XDS | 46.50% | 8.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 6.9 | - | - | - | |||||
| 7gjs | 2.00 Å | 2023-11-08 | X-ray | QQO | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.39% | 27.86% | 12.80% | (2R)-N-{(2R)-2-(3,4-dichlorophenyl)-1-[(isoquinolin-4-yl)amino]-1-oxopropan-2-yl}-4-(propan-2-yl)morpholine-2-carboxamide | (2R)-N-{(2R)-2-(3,4-dichlorophenyl)-1-[(isoquinolin-4-yl)amino]-1-oxopropan-2-yl}-4-(propan-2-yl)morpholine-2-carboxamide | XDS | 64.20% | 10.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 14.8 | - | - | - | |||||
| 7gjt | 2.12 Å | 2023-11-08 | X-ray | QQU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.86% | 28.57% | 15.60% | (4S)-6-chloro-N-(isoquinolin-4-yl)-4-{2-oxo-2-[(2R)-2-(1H-pyrazol-4-yl)piperidin-1-yl]ethyl}-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-4-{2-oxo-2-[(2R)-2-(1H-pyrazol-4-yl)piperidin-1-yl]ethyl}-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 56.40% | 9.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 11.7 | - | - | - | |||||
| 7gju | 1.94 Å | 2023-11-08 | X-ray | QR5 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.06% | 27.31% | 16.50% | (2R)-2-[2-(3-cyclopropyl-2-oxoimidazolidin-1-yl)acetamido]-2-(3,4-dichlorophenyl)-N-(isoquinolin-4-yl)propanamide | (2R)-2-[2-(3-cyclopropyl-2-oxoimidazolidin-1-yl)acetamido]-2-(3,4-dichlorophenyl)-N-(isoquinolin-4-yl)propanamide | XDS | 51.00% | 8.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 17.7 | - | - | - | |||||
| 7gjv | 2.09 Å | 2023-11-08 | X-ray | QP6 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.25% | 27.49% | 13.80% | (4S)-6,8-difluoro-N-(isoquinolin-4-yl)-4-methoxy-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4S)-6,8-difluoro-N-(isoquinolin-4-yl)-4-methoxy-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 57.20% | 10.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 16.8 | - | - | - | |||||
| 7gjw | 2.46 Å | 2023-11-08 | X-ray | QR9 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 24.02% | 30.33% | 36.00% | (4S)-4-{2-[(1R,4R)-5-acetyl-2,5-diazabicyclo[2.2.1]heptan-2-yl]-2-oxoethyl}-6-chloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4S)-4-{2-[(1R,4R)-5-acetyl-2,5-diazabicyclo[2.2.1]heptan-2-yl]-2-oxoethyl}-6-chloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 68.60% | 4.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 6.8 | - | - | - | |||||
| 7gjx | 2.06 Å | 2023-11-08 | X-ray | QNU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 24.18% | 29.13% | 18.70% | (4S)-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 59.20% | 6.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 9.5 | - | - | - | |||||
| 7gjy | 2.18 Å | 2023-11-08 | X-ray | QRS | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.74% | 28.42% | 34.10% | (3S)-N-{(2R)-2-(3,4-dichlorophenyl)-1-[(isoquinolin-4-yl)amino]-1-oxopropan-2-yl}-1-methylpyrrolidine-3-carboxamide | (3S)-N-{(2R)-2-(3,4-dichlorophenyl)-1-[(isoquinolin-4-yl)amino]-1-oxopropan-2-yl}-1-methylpyrrolidine-3-carboxamide | XDS | 100.00% | 4.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 12.2 | - | - | - | |||||
| 7gjz | 1.65 Å | 2023-11-08 | X-ray | QRF | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.05% | 25.89% | 10.20% | 2-{3-chloro-5-[4-(furan-2-carbonyl)piperazin-1-yl]phenyl}-N-(isoquinolin-4-yl)acetamide | 2-{3-chloro-5-[4-(furan-2-carbonyl)piperazin-1-yl]phenyl}-N-(isoquinolin-4-yl)acetamide | XDS | 52.70% | 10.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 27.6 | - | - | - | |||||
| 7gk0 | 1.96 Å | 2023-11-08 | X-ray | QS3 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 24.14% | 28.73% | 16.40% | (1'M,4S)-6-chloro-1'-(isoquinolin-4-yl)-3'-methyl-2,3-dihydrospiro[[1]benzopyran-4,4'-imidazolidine]-2',5'-dione | (1'M,4S)-6-chloro-1'-(isoquinolin-4-yl)-3'-methyl-2,3-dihydrospiro[[1]benzopyran-4,4'-imidazolidine]-2',5'-dione | XDS | 54.00% | 8.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 10.9 | - | - | - | |||||
| 7gk1 | 1.94 Å | 2023-11-08 | X-ray | QSF | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.93% | 26.89% | 58.60% | (3S)-5-chloro-N-(isoquinolin-4-yl)-3-methyl-2-oxo-2,3-dihydro-1H-indole-3-carboxamide | (3S)-5-chloro-N-(isoquinolin-4-yl)-3-methyl-2-oxo-2,3-dihydro-1H-indole-3-carboxamide | XDS | 48.00% | 4.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 20.3 | - | - | - | |||||
| 7gk2 | 2.01 Å | 2023-11-08 | X-ray | QSX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 24.57% | 28.74% | 20.10% | (4S)-6-chloro-N-(isoquinolin-4-yl)-4-{2-[(1S,4S)-5-methyl-2,5-diazabicyclo[2.2.1]heptan-2-yl]-2-oxoethyl}-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-4-{2-[(1S,4S)-5-methyl-2,5-diazabicyclo[2.2.1]heptan-2-yl]-2-oxoethyl}-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 57.30% | 6.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 10.9 | - | - | - | |||||
| 7gk3 | 2.19 Å | 2023-11-08 | X-ray | QT3 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.69% | 28.27% | 22.90% | (1R)-7-chloro-N-(isoquinolin-4-yl)-2-methyl-1,2,3,4-tetrahydroisoquinoline-1-carboxamide | (1R)-7-chloro-N-(isoquinolin-4-yl)-2-methyl-1,2,3,4-tetrahydroisoquinoline-1-carboxamide | XDS | 51.80% | 7.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 12.9 | - | - | - | |||||
| 7gk4 | 2.23 Å | 2023-11-08 | X-ray | QTC | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.26% | 28.43% | 42.50% | 2-(3-chloro-5-ethylphenyl)-N-(isoquinolin-4-yl)acetamide | 2-(3-chloro-5-ethylphenyl)-N-(isoquinolin-4-yl)acetamide | XDS | 100.00% | 4.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 12.2 | - | - | - | |||||
| 7gk5 | 1.97 Å | 2023-11-08 | X-ray | QC3 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.92% | 26.79% | 15.60% | (3S)-3-(3-chlorophenyl)-1-(isoquinolin-4-yl)pyrrolidin-2-one | (3S)-3-(3-chlorophenyl)-1-(isoquinolin-4-yl)pyrrolidin-2-one | XDS | 66.20% | 8.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 21.1 | - | - | - | |||||
| 7gk6 | 1.98 Å | 2023-11-08 | X-ray | QTL | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 24.79% | 28.84% | 15.30% | (1R)-7-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-1-carboxamide | (1R)-7-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-1-carboxamide | XDS | 53.70% | 8.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 10.5 | - | - | - | |||||
| 7gk7 | 2.18 Å | 2023-11-08 | X-ray | QU9 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 24.77% | 29.18% | 21.90% | (4S)-6-chloro-4-(2-{(2R)-2-[(1H-imidazol-1-yl)methyl]pyrrolidin-1-yl}-2-oxoethyl)-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4S)-6-chloro-4-(2-{(2R)-2-[(1H-imidazol-1-yl)methyl]pyrrolidin-1-yl}-2-oxoethyl)-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 59.40% | 7.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 9.4 | - | - | - | |||||
| 7gk8 | 2.20 Å | 2023-11-08 | X-ray | QUQ | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.91% | 28.90% | 45.60% | (4S)-2-acetyl-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-2-acetyl-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 99.90% | 5.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 10.2 | - | - | - | |||||
| 7gk9 | 2.02 Å | 2023-11-08 | X-ray | QV0 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.25% | 27.75% | 35.90% | (3'R)-6-chloro-1'-(isoquinolin-4-yl)-2,3-dihydrospiro[[1]benzopyran-4,3'-piperidin]-2'-one | (3'R)-6-chloro-1'-(isoquinolin-4-yl)-2,3-dihydrospiro[[1]benzopyran-4,3'-piperidin]-2'-one | XDS | 99.70% | 4.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 15.4 | - | - | - | |||||
| 7gka | 2.06 Å | 2023-11-08 | X-ray | QV9 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 24.34% | 28.27% | 18.20% | (3R)-3-(3-chlorophenyl)-1-(isoquinolin-4-yl)pyrrolidin-2-one | (3R)-3-(3-chlorophenyl)-1-(isoquinolin-4-yl)pyrrolidin-2-one | XDS | 59.20% | 7.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 12.9 | - | - | - | |||||
| 7gkb | 1.96 Å | 2023-11-08 | X-ray | QVG | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.94% | 28.45% | 17.10% | (4R)-6-chloro-7-fluoro-N-(6-fluoroisoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-7-fluoro-N-(6-fluoroisoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 61.20% | 7.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 12.1 | - | - | - | |||||
| 7gkc | 1.94 Å | 2023-11-08 | X-ray | QVJ | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.03% | 26.77% | 12.50% | (4R)-6-chloro-N-(isoquinolin-4-yl)-4-{[2-(methylamino)-2-oxoethoxy]methyl}-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-(isoquinolin-4-yl)-4-{[2-(methylamino)-2-oxoethoxy]methyl}-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 55.60% | 10.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 21.1 | - | - | - | |||||
| 7gkd | 2.24 Å | 2023-11-08 | X-ray | QVU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 24.44% | 29.87% | 22.10% | (4R)-6,7-dichloro-N-(2,7-naphthyridin-4-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide | (4R)-6,7-dichloro-N-(2,7-naphthyridin-4-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide | XDS | 50.90% | 7.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 7.6 | - | - | - | |||||
| 7gke | 1.97 Å | 2023-11-08 | X-ray | QW1 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.94% | 28.14% | 16.70% | (4S)-6-chloro-N~4~-(isoquinolin-4-yl)-3,4-dihydroisoquinoline-2,4(1H)-dicarboxamide | (4S)-6-chloro-N~4~-(isoquinolin-4-yl)-3,4-dihydroisoquinoline-2,4(1H)-dicarboxamide | XDS | 52.90% | 8.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 13.4 | - | - | - | |||||
| 7gkf | 1.85 Å | 2023-11-08 | X-ray | QWL | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.10% | 26.32% | 16.10% | (4R)-6,7-dichloro-N-(6-fluoroisoquinolin-4-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide | (4R)-6,7-dichloro-N-(6-fluoroisoquinolin-4-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide | XDS | 59.20% | 8.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 24.2 | - | - | - | |||||
| 7gkg | 2.03 Å | 2023-11-08 | X-ray | QWU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.13% | 27.89% | 14.20% | (3S)-5-chloro-N-(isoquinolin-4-yl)-3-methyl-2,3-dihydro-1H-indole-3-carboxamide | (3S)-5-chloro-N-(isoquinolin-4-yl)-3-methyl-2,3-dihydro-1H-indole-3-carboxamide | XDS | 51.50% | 9.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 14.7 | - | - | - | |||||
| 7gkh | 1.99 Å | 2023-11-08 | X-ray | QX3 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.91% | 28.21% | 14.90% | (4R)-6,7-dichloro-N-(4-cyclopropylpyridin-3-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide | (4R)-6,7-dichloro-N-(4-cyclopropylpyridin-3-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide | XDS | 62.20% | 9.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 13.1 | - | - | - | |||||
| 7gki | 2.00 Å | 2023-11-08 | X-ray | QX9 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 24.68% | 29.11% | 26.40% | 2-(3-chlorophenyl)-N-(6-methylisoquinolin-4-yl)acetamide | 2-(3-chlorophenyl)-N-(6-methylisoquinolin-4-yl)acetamide | XDS | 72.90% | 6.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 9.6 | - | - | - | |||||
| 7gkj | 1.76 Å | 2023-11-08 | X-ray | QXI | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.38% | 27.42% | 9.70% | (4S)-6-chloro-N-(isoquinolin-4-yl)-4-{2-[(4R,8S)-8-methyl-5,6-dihydro[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl]-2-oxoethyl}-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-4-{2-[(4R,8S)-8-methyl-5,6-dihydro[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl]-2-oxoethyl}-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 58.80% | 11.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 17.1 | - | - | - | |||||
| 7gkk | 1.80 Å | 2023-11-08 | X-ray | QXR | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.85% | 27.65% | 9.50% | N-(4-tert-butoxypyridin-3-yl)-2-(3-chlorophenyl)acetamide | N-(4-tert-butoxypyridin-3-yl)-2-(3-chlorophenyl)acetamide | XDS | 58.50% | 11.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 15.9 | - | - | - | |||||
| 7gkl | 1.93 Å | 2023-11-08 | X-ray | QXX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.92% | 27.67% | 12.50% | 2-(3-chlorophenyl)-N-[4-(trifluoromethyl)pyridin-3-yl]acetamide | 2-(3-chlorophenyl)-N-[4-(trifluoromethyl)pyridin-3-yl]acetamide | XDS | 55.00% | 10.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 15.8 | - | - | - | |||||
| 7gkm | 1.95 Å | 2023-11-08 | X-ray | QY6 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.15% | 27.98% | 12.80% | (3R)-6'-chloro-1-(isoquinolin-4-yl)-2',3'-dihydro-1'H-spiro[piperidine-3,4'-quinolin]-2-one | (3R)-6'-chloro-1-(isoquinolin-4-yl)-2',3'-dihydro-1'H-spiro[piperidine-3,4'-quinolin]-2-one | XDS | 55.90% | 9.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 14.2 | - | - | - | |||||
| 7gkn | 1.92 Å | 2023-11-08 | X-ray | QYI | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.79% | 27.42% | 15.70% | (4S)-6-chloro-N-(6-fluoroisoquinolin-4-yl)-4-methoxy-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4S)-6-chloro-N-(6-fluoroisoquinolin-4-yl)-4-methoxy-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 68.30% | 8.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 17.1 | - | - | - | |||||
| 7gko | 2.05 Å | 2023-11-08 | X-ray | QYN | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.92% | 27.37% | 14.20% | (4S)-6-chloro-2-(1H-imidazole-2-sulfonyl)-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-2-(1H-imidazole-2-sulfonyl)-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 52.20% | 9.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 17.5 | - | - | - | |||||
| 7gkp | 1.79 Å | 2023-11-08 | X-ray | QYR | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.82% | 26.32% | 11.60% | (4S)-6-chloro-2-(cyclopropanesulfonyl)-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-2-(cyclopropanesulfonyl)-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 60.30% | 9.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 24.2 | - | - | - | |||||
| 7gkq | 2.07 Å | 2023-11-08 | X-ray | QZ0 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.40% | 27.20% | 14.70% | (4S)-6-chloro-N~4~-(isoquinolin-4-yl)-N~2~,N~2~-dimethyl-3,4-dihydroisoquinoline-2,4(1H)-dicarboxamide | (4S)-6-chloro-N~4~-(isoquinolin-4-yl)-N~2~,N~2~-dimethyl-3,4-dihydroisoquinoline-2,4(1H)-dicarboxamide | XDS | 56.70% | 9.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 18.3 | - | - | - | |||||
| 7gkr | 2.02 Å | 2023-11-08 | X-ray | QZC | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.35% | 26.53% | 14.50% | (4S)-6-chloro-2-(dimethylsulfamoyl)-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-2-(dimethylsulfamoyl)-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 53.20% | 8.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 22.7 | - | - | - | |||||
| 7gks | 1.81 Å | 2023-11-08 | X-ray | QZL | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.04% | 25.00% | 12.20% | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[(3R)-3-methylpyrrolidine-1-sulfonyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[(3R)-3-methylpyrrolidine-1-sulfonyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 62.00% | 10.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 35.1 | - | - | - | |||||
| 7gkt | 1.84 Å | 2023-11-08 | X-ray | QZU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.95% | 26.04% | 13.80% | (4R)-6-chloro-N-(isoquinolin-4-yl)-4-[(prop-2-enamido)methyl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-(isoquinolin-4-yl)-4-[(prop-2-enamido)methyl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 61.90% | 9.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 26.2 | - | - | - | |||||
| 7gku | 1.87 Å | 2023-11-08 | X-ray | R08 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.53% | 25.77% | 15.00% | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-(1-methyl-1H-pyrazole-5-carbonyl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-(1-methyl-1H-pyrazole-5-carbonyl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 56.10% | 6.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 28.6 | - | - | - | |||||
| 7gkv | 1.88 Å | 2023-11-08 | X-ray | R0F | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.85% | 26.44% | 15.10% | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[2-(methylamino)-2-oxoethyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[2-(methylamino)-2-oxoethyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 55.40% | 9.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 23.4 | - | - | - | |||||
| 7gkw | 1.91 Å | 2023-11-08 | X-ray | R0Q | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.32% | 25.81% | 15.10% | methyl ({(4R)-6-chloro-4-[(isoquinolin-4-yl)carbamoyl]-3,4-dihydro-2H-1-benzopyran-4-yl}methyl)carbamate | methyl ({(4R)-6-chloro-4-[(isoquinolin-4-yl)carbamoyl]-3,4-dihydro-2H-1-benzopyran-4-yl}methyl)carbamate | XDS | 72.50% | 9.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 28.1 | - | - | - | |||||
| 7gkx | 1.87 Å | 2023-11-08 | X-ray | R1I | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.40% | 28.20% | 17.60% | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-(2-methoxyethyl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-(2-methoxyethyl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 51.80% | 6.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 13.1 | - | - | - | |||||
| 7gky | 1.97 Å | 2023-11-08 | X-ray | R1U | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.01% | 26.25% | 15.30% | 2-(3-chlorophenyl)-N-(1-methyl-1H-pyrazolo[4,3-c]pyridin-7-yl)acetamide | 2-(3-chlorophenyl)-N-(1-methyl-1H-pyrazolo[4,3-c]pyridin-7-yl)acetamide | XDS | 57.40% | 9.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 24.8 | - | - | - | |||||
| 7gkz | 1.81 Å | 2023-11-08 | X-ray | R2L | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.80% | 25.76% | 17.50% | (4R)-6-chloro-N-[4-methyl-5-(methylamino)pyridin-3-yl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4R)-6-chloro-N-[4-methyl-5-(methylamino)pyridin-3-yl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 61.10% | 8.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 28.6 | - | - | - | |||||
| 7gl0 | 1.82 Å | 2023-11-08 | X-ray | R2X | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.57% | 25.72% | 12.80% | (2S,5R)-N-{(1R)-1-(3-chlorophenyl)-2-[(isoquinolin-4-yl)amino]-2-oxoethyl}-5-(pyrrolidine-1-carbonyl)oxolane-2-carboxamide (non-preferred name) | (2S,5R)-N-{(1R)-1-(3-chlorophenyl)-2-[(isoquinolin-4-yl)amino]-2-oxoethyl}-5-(pyrrolidine-1-carbonyl)oxolane-2-carboxamide (non-preferred name) | XDS | 67.40% | 10.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 28.9 | - | - | - | |||||
| 7gl1 | 1.71 Å | 2023-11-08 | X-ray | R3I | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.13% | 23.93% | 9.00% | (3S)-5-chloro-1'-(6-fluoroisoquinolin-4-yl)-2H-spiro[[1]benzofuran-3,3'-pyrrolidin]-2'-one | (3S)-5-chloro-1'-(6-fluoroisoquinolin-4-yl)-2H-spiro[[1]benzofuran-3,3'-pyrrolidin]-2'-one | XDS | 70.20% | 12.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 45.5 | - | - | - | |||||
| 7gl2 | 1.81 Å | 2023-11-08 | X-ray | R43 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.91% | 25.88% | 10.50% | (4S)-6-chloro-2-(3-cyanoazetidine-1-sulfonyl)-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-2-(3-cyanoazetidine-1-sulfonyl)-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 70.40% | 10.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 27.7 | - | - | - | |||||
| 7gl3 | 1.75 Å | 2023-11-08 | X-ray | R4X | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.06% | 24.21% | 10.60% | (4S)-2-(azetidine-1-sulfonyl)-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-2-(azetidine-1-sulfonyl)-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 71.00% | 10.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 42.8 | - | - | - | |||||
| 7gl4 | 1.87 Å | 2023-11-08 | X-ray | R5H | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.84% | 25.44% | 10.70% | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-(3-methoxyazetidine-1-sulfonyl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-(3-methoxyazetidine-1-sulfonyl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 70.90% | 10.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 31.3 | - | - | - | |||||
| 7gl5 | 1.68 Å | 2023-11-08 | X-ray | R5O | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.99% | 25.74% | 11.00% | (4S)-6-chloro-N-{6-[(methanesulfonyl)amino]isoquinolin-4-yl}-4-methoxy-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4S)-6-chloro-N-{6-[(methanesulfonyl)amino]isoquinolin-4-yl}-4-methoxy-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 61.70% | 10.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 28.8 | - | - | - | |||||
| 7gl6 | 1.72 Å | 2023-11-08 | X-ray | R66 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.06% | 24.25% | 10.10% | (3R)-3-(3-chlorophenyl)-3-hydroxy-1-(isoquinolin-4-yl)pyrrolidin-2-one | (3R)-3-(3-chlorophenyl)-3-hydroxy-1-(isoquinolin-4-yl)pyrrolidin-2-one | XDS | 70.90% | 11.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 42.5 | - | - | - | |||||
| 7gl7 | 1.85 Å | 2023-11-08 | X-ray | R6L | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.61% | 24.85% | 13.80% | (4S)-6-chloro-4-methoxy-N-[7-(methylsulfamoyl)isoquinolin-4-yl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4S)-6-chloro-4-methoxy-N-[7-(methylsulfamoyl)isoquinolin-4-yl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 63.30% | 9.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 36.7 | - | - | - | |||||
| 7gl8 | 1.64 Å | 2023-11-08 | X-ray | KG9 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.68% | 24.20% | 8.60% | (4S)-6-chloro-2-[(1-cyanocyclopropyl)methanesulfonyl]-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-2-[(1-cyanocyclopropyl)methanesulfonyl]-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 70.70% | 11.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 42.9 | - | - | - | |||||
| 7gl9 | 1.94 Å | 2023-11-08 | X-ray | QM3 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 24.04% | 28.88% | 17.50% | (1'P,3'S)-6-chloro-1'-(isoquinolin-4-yl)-2,3-dihydrospiro[[1]benzopyran-4,3'-pyrrolidine]-2',5'-dione | (1'P,3'S)-6-chloro-1'-(isoquinolin-4-yl)-2,3-dihydrospiro[[1]benzopyran-4,3'-pyrrolidine]-2',5'-dione | XDS | 52.50% | 8.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 10.4 | - | - | - | |||||
| 7gla | 1.68 Å | 2023-11-08 | X-ray | R7F | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.10% | 24.52% | 10.20% | (4R)-6-chloro-N-[6-(2-hydroxypropan-2-yl)isoquinolin-4-yl]-1,2,3,4-tetrahydroquinoline-4-carboxamide | (4R)-6-chloro-N-[6-(2-hydroxypropan-2-yl)isoquinolin-4-yl]-1,2,3,4-tetrahydroquinoline-4-carboxamide | XDS | 58.40% | 10.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 39.8 | - | - | - | |||||
| 7glb | 1.95 Å | 2023-11-08 | X-ray | R76 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.29% | 24.86% | 12.50% | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[2-(methylamino)-2-oxoethyl]-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[2-(methylamino)-2-oxoethyl]-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 65.40% | 13.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 36.6 | - | - | - | |||||
| 7glc | 1.95 Å | 2023-11-08 | X-ray | R87 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.95% | 27.06% | 15.30% | (4S)-6-chloro-N-(7-fluoroisoquinolin-4-yl)-4-methoxy-3,4-dihydro-2H-1-benzopyran-4-carboxamide | (4S)-6-chloro-N-(7-fluoroisoquinolin-4-yl)-4-methoxy-3,4-dihydro-2H-1-benzopyran-4-carboxamide | XDS | 67.40% | 7.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 19.2 | - | - | - | |||||
| 7gld | 1.78 Å | 2023-11-08 | X-ray | R8I | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.98% | 24.83% | 14.10% | (4S)-6-chloro-2-{2-[(cyanomethyl)amino]-2-oxoethyl}-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-2-{2-[(cyanomethyl)amino]-2-oxoethyl}-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 58.20% | 8.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 36.8 | - | - | - | |||||
| 7gle | 1.98 Å | 2023-11-08 | X-ray | R8O | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.70% | 25.88% | 12.90% | (4S)-6-chloro-2-[2-(cyclopropylamino)-2-oxoethyl]-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-2-[2-(cyclopropylamino)-2-oxoethyl]-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 64.30% | 9.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 27.7 | - | - | - | |||||
| 7glf | 1.67 Å | 2023-11-08 | X-ray | R8X | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.22% | 23.97% | 11.20% | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[(2-methoxyethyl)(methyl)sulfamoyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[(2-methoxyethyl)(methyl)sulfamoyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 69.20% | 9.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 45.2 | - | - | - | |||||
| 7glg | 1.67 Å | 2023-11-08 | X-ray | R95 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.27% | 23.85% | 11.10% | (4S)-6-chloro-2-[(cyanomethyl)(methyl)sulfamoyl]-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-2-[(cyanomethyl)(methyl)sulfamoyl]-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 58.20% | 9.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 46.4 | - | - | - | |||||
| 7glh | 1.91 Å | 2023-11-08 | X-ray | R9E | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.18% | 25.71% | 13.70% | 2-(3-chlorophenyl)-N-(7-fluoro-6-methoxyisoquinolin-4-yl)acetamide | 2-(3-chlorophenyl)-N-(7-fluoro-6-methoxyisoquinolin-4-yl)acetamide | XDS | 64.30% | 10.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 28.9 | - | - | - | |||||
| 7gli | 1.84 Å | 2023-11-08 | X-ray | R9I | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.43% | 25.57% | 17.00% | methyl N-[(4S)-6-chloro-4-[(isoquinolin-4-yl)carbamoyl]-3,4-dihydroisoquinoline-2(1H)-sulfonyl]-N-methylglycinate | methyl N-[(4S)-6-chloro-4-[(isoquinolin-4-yl)carbamoyl]-3,4-dihydroisoquinoline-2(1H)-sulfonyl]-N-methylglycinate | XDS | 55.60% | 8.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 30.3 | - | - | - | |||||
| 7glj | 1.81 Å | 2023-11-08 | X-ray | R9R | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.54% | 24.71% | 12.00% | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[(3-methyl-1,1-dioxo-1lambda~6~-thietan-3-yl)methanesulfonyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[(3-methyl-1,1-dioxo-1lambda~6~-thietan-3-yl)methanesulfonyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 55.40% | 10.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 38.0 | - | - | - | |||||
| 7glk | 1.74 Å | 2023-11-08 | X-ray | R9Z | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.86% | 24.00% | 9.40% | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[(1-methoxycyclopropyl)methanesulfonyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[(1-methoxycyclopropyl)methanesulfonyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 72.80% | 10.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 44.7 | - | - | - | |||||
| 7gll | 1.91 Å | 2023-11-08 | X-ray | RAQ | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.63% | 24.49% | 10.50% | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[(oxan-4-yl)methanesulfonyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[(oxan-4-yl)methanesulfonyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 74.80% | 10.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 40.4 | - | - | - | |||||
| 7glm | 1.91 Å | 2023-11-08 | X-ray | RBM | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.45% | 25.97% | 16.40% | (4S)-6-chloro-2-[(2-cyanoethyl)(methyl)sulfamoyl]-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-2-[(2-cyanoethyl)(methyl)sulfamoyl]-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 54.80% | 7.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 26.9 | - | - | - | |||||
| 7gln | 1.84 Å | 2023-11-08 | X-ray | RBX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.21% | 24.00% | 11.70% | (4S)-6-chloro-2-[ethyl(methyl)sulfamoyl]-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-2-[ethyl(methyl)sulfamoyl]-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 68.20% | 9.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 44.7 | - | - | - | |||||
| 7glo | 1.93 Å | 2023-11-08 | X-ray | L6D | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.79% | 24.64% | 14.30% | N-(1H-benzimidazol-1-yl)-2-(3-chlorophenyl)acetamide | N-(1H-benzimidazol-1-yl)-2-(3-chlorophenyl)acetamide | XDS | 60.20% | 9.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 38.7 | - | - | - | |||||
| 7glp | 1.92 Å | 2023-11-08 | X-ray | 860 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.57% | 25.02% | 11.40% | 2-(3-chlorophenyl)-N-(isoquinolin-4-yl)acetamide | 2-(3-chlorophenyl)-N-(isoquinolin-4-yl)acetamide | XDS | 80.70% | 10.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 34.9 | - | - | - | |||||
| 7glq | 2.06 Å | 2023-11-08 | X-ray | NM0 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.85% | 24.38% | 14.30% | 2-(3-chlorophenyl)-N-[(4R)-imidazo[1,2-a]pyridin-3-yl]acetamide | 2-(3-chlorophenyl)-N-[(4R)-imidazo[1,2-a]pyridin-3-yl]acetamide | XDS | 82.10% | 9.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 41.4 | - | - | - | |||||
| 7glr | 1.79 Å | 2023-11-08 | X-ray | O0X | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.96% | 24.48% | 10.00% | 2-(3-chlorophenyl)-N-(1,7-naphthyridin-5-yl)acetamide | 2-(3-chlorophenyl)-N-(1,7-naphthyridin-5-yl)acetamide | XDS | 60.90% | 10.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 40.4 | - | - | - | |||||
| 7gls | 1.73 Å | 2023-11-08 | X-ray | OE6 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.70% | 26.13% | 13.10% | N-(1H-benzotriazol-1-yl)-2-(3-chlorophenyl)acetamide | N-(1H-benzotriazol-1-yl)-2-(3-chlorophenyl)acetamide | XDS | 56.90% | 9.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 25.6 | - | - | - | |||||
| 7glt | 1.86 Å | 2023-11-08 | X-ray | RC9 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.62% | 24.09% | 12.40% | (4S)-6-chloro-2-[(1-cyanocyclobutyl)methanesulfonyl]-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-2-[(1-cyanocyclobutyl)methanesulfonyl]-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 72.00% | 9.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 44.1 | - | - | - | |||||
| 7glu | 1.63 Å | 2023-11-08 | X-ray | RD5 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.15% | 24.49% | 9.80% | 2-(5-chloro-2-{[(methanesulfonyl)amino]methyl}phenyl)-N-(isoquinolin-4-yl)acetamide | 2-(5-chloro-2-{[(methanesulfonyl)amino]methyl}phenyl)-N-(isoquinolin-4-yl)acetamide | XDS | 59.30% | 11.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 40.4 | - | - | - | |||||
| 7glv | 1.72 Å | 2023-11-08 | X-ray | R76 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.14% | 24.81% | 13.90% | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[2-(methylamino)-2-oxoethyl]-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[2-(methylamino)-2-oxoethyl]-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 55.50% | 9.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 37.0 | - | - | - | |||||
| 7glw | 1.59 Å | 2023-11-08 | X-ray | RDK | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.89% | 23.66% | 7.90% | (4S)-6-chloro-N-(isoquinolin-4-yl)-4-methyl-1,1-dioxo-1,2,3,4-tetrahydro-1lambda~6~,2-benzothiazine-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-4-methyl-1,1-dioxo-1,2,3,4-tetrahydro-1lambda~6~,2-benzothiazine-4-carboxamide | XDS | 62.40% | 12.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 48.3 | - | - | - | |||||
| 7glx | 1.49 Å | 2023-11-08 | X-ray | RDQ | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.17% | 24.14% | 11.20% | (4S)-6-chloro-N-{6-[(methanesulfonyl)amino]isoquinolin-4-yl}-2-[2-(methylamino)-2-oxoethyl]-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-{6-[(methanesulfonyl)amino]isoquinolin-4-yl}-2-[2-(methylamino)-2-oxoethyl]-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 65.30% | 8.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 43.5 | - | - | - | |||||
| 7gly | 1.80 Å | 2023-11-08 | X-ray | RDX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.40% | 24.29% | 12.10% | (4S)-6-chloro-2-[(2-hydroxyethyl)(methyl)sulfamoyl]-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-2-[(2-hydroxyethyl)(methyl)sulfamoyl]-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 61.20% | 11.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 42.2 | - | - | - | |||||
| 7glz | 1.68 Å | 2023-11-08 | X-ray | REU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.74% | 25.37% | 13.60% | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[(2S)-1-(methylamino)-1-oxopropan-2-yl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[(2S)-1-(methylamino)-1-oxopropan-2-yl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 61.70% | 9.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 32.0 | - | - | - | |||||
| 7gm0 | 1.76 Å | 2023-11-08 | X-ray | RFF | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.71% | 25.09% | 12.90% | (4S)-6-chloro-2-[(1-cyanocyclopropyl)methanesulfonyl]-N-(1-methyl-1H-pyrazolo[4,3-c]pyridin-7-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-2-[(1-cyanocyclopropyl)methanesulfonyl]-N-(1-methyl-1H-pyrazolo[4,3-c]pyridin-7-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 62.00% | 8.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 34.5 | - | - | - | |||||
| 7gm1 | 1.94 Å | 2023-11-08 | X-ray | RFR | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.28% | 26.08% | 12.50% | (4S)-6-chloro-N-{6-[(methanesulfonyl)amino]isoquinolin-4-yl}-2-[2-(methylamino)-2-oxoethyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-{6-[(methanesulfonyl)amino]isoquinolin-4-yl}-2-[2-(methylamino)-2-oxoethyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 61.30% | 9.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 26.1 | - | - | - | |||||
| 7gm2 | 1.89 Å | 2023-11-08 | X-ray | RG3 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.96% | 24.41% | 15.60% | (4S)-6-chloro-N-[7-(methanesulfonyl)isoquinolin-4-yl]-2-[2-(methylamino)-2-oxoethyl]-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-[7-(methanesulfonyl)isoquinolin-4-yl]-2-[2-(methylamino)-2-oxoethyl]-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 57.20% | 7.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 40.9 | - | - | - | |||||
| 7gm3 | 2.02 Å | 2023-11-08 | X-ray | RG9 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.03% | 25.42% | 16.10% | 2-(3-chlorophenyl)-N-[7-(2-hydroxypropan-2-yl)isoquinolin-4-yl]acetamide | 2-(3-chlorophenyl)-N-[7-(2-hydroxypropan-2-yl)isoquinolin-4-yl]acetamide | XDS | 54.20% | 9.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 31.4 | - | - | - | |||||
| 7gm4 | 1.75 Å | 2023-11-08 | X-ray | RGQ | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.03% | 26.35% | 13.80% | (4S)-6-chloro-N-(7-fluoroisoquinolin-4-yl)-2-[2-(methylamino)-2-oxoethyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-(7-fluoroisoquinolin-4-yl)-2-[2-(methylamino)-2-oxoethyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 54.90% | 7.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 24.1 | - | - | - | |||||
| 7gm5 | 1.71 Å | 2023-11-08 | X-ray | RGX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.78% | 24.68% | 9.10% | (4S)-6-chloro-2-(cyclopropylsulfamoyl)-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-2-(cyclopropylsulfamoyl)-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 67.70% | 11.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 38.5 | - | - | - | |||||
| 7gm6 | 1.87 Å | 2023-11-08 | X-ray | RHI | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.63% | 25.45% | 14.20% | (4S)-6-chloro-N-[7-(methanesulfonyl)isoquinolin-4-yl]-2-[2-(methylamino)-2-oxoethyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-[7-(methanesulfonyl)isoquinolin-4-yl]-2-[2-(methylamino)-2-oxoethyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 60.50% | 8.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 31.2 | - | - | - | |||||
| 7gm7 | 1.84 Å | 2023-11-08 | X-ray | RI1 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.45% | 25.56% | 14.50% | (4S)-6-chloro-N-(isoquinolin-4-yl)-1,1-dioxo-1,2,3,4-tetrahydro-1lambda~6~,2-benzothiazine-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-1,1-dioxo-1,2,3,4-tetrahydro-1lambda~6~,2-benzothiazine-4-carboxamide | XDS | 64.60% | 9.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 30.3 | - | - | - | |||||
| 7gm8 | 1.73 Å | 2023-11-08 | X-ray | RI6 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.56% | 24.39% | 12.10% | (4S)-6-chloro-N-(7-chloroisoquinolin-4-yl)-2-[2-(methylamino)-2-oxoethyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-(7-chloroisoquinolin-4-yl)-2-[2-(methylamino)-2-oxoethyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 57.60% | 9.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 41.3 | - | - | - | |||||
| 7gm9 | 1.83 Å | 2023-11-08 | X-ray | RIJ | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.40% | 25.10% | 5.80% | 2-[(1'P,3'S)-6-chloro-1'-(isoquinolin-4-yl)-2',5'-dioxo-1H-spiro[isoquinoline-4,3'-pyrrolidin]-2(3H)-yl]-N-methylacetamide | 2-[(1'P,3'S)-6-chloro-1'-(isoquinolin-4-yl)-2',5'-dioxo-1H-spiro[isoquinoline-4,3'-pyrrolidin]-2(3H)-yl]-N-methylacetamide | XDS | 65.30% | 15.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 34.2 | - | - | - | |||||
| 7gma | 1.86 Å | 2023-11-08 | X-ray | R2X | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.69% | 24.83% | 8.20% | (2S,5R)-N-{(1R)-1-(3-chlorophenyl)-2-[(isoquinolin-4-yl)amino]-2-oxoethyl}-5-(pyrrolidine-1-carbonyl)oxolane-2-carboxamide (non-preferred name) | (2S,5R)-N-{(1R)-1-(3-chlorophenyl)-2-[(isoquinolin-4-yl)amino]-2-oxoethyl}-5-(pyrrolidine-1-carbonyl)oxolane-2-carboxamide (non-preferred name) | XDS | 61.50% | 12.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 36.8 | - | - | - | |||||
| 7gmb | 2.21 Å | 2023-11-08 | X-ray | RIU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.75% | 24.60% | 7.70% | 2-(3-chlorophenyl)-N-{6-[2-(dimethylamino)ethoxy]isoquinolin-4-yl}acetamide | 2-(3-chlorophenyl)-N-{6-[2-(dimethylamino)ethoxy]isoquinolin-4-yl}acetamide | XDS | 69.00% | 14.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 39.0 | - | - | - | |||||
| 7gmc | 1.82 Å | 2023-11-08 | X-ray | RIY | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.78% | 23.80% | 6.40% | (4S)-6-chloro-2-(ethylsulfamoyl)-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-2-(ethylsulfamoyl)-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 68.60% | 14.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 46.8 | - | - | - | |||||
| 7gmd | 1.94 Å | 2023-11-08 | X-ray | R2X | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.54% | 24.27% | 8.60% | (2S,5R)-N-{(1R)-1-(3-chlorophenyl)-2-[(isoquinolin-4-yl)amino]-2-oxoethyl}-5-(pyrrolidine-1-carbonyl)oxolane-2-carboxamide (non-preferred name) | (2S,5R)-N-{(1R)-1-(3-chlorophenyl)-2-[(isoquinolin-4-yl)amino]-2-oxoethyl}-5-(pyrrolidine-1-carbonyl)oxolane-2-carboxamide (non-preferred name) | XDS | 71.30% | 12.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 42.4 | - | - | - | |||||
| 7gme | 1.85 Å | 2023-11-08 | X-ray | RJ3 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.04% | 24.39% | 9.80% | 4-[2-(3-chlorophenyl)acetamido]isoquinoline-7-carboxylic acid | 4-[2-(3-chlorophenyl)acetamido]isoquinoline-7-carboxylic acid | XDS | 62.80% | 9.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 41.3 | - | - | - | |||||
| 7gmf | 1.80 Å | 2023-11-08 | X-ray | RJF | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.61% | 23.74% | 5.50% | (4S)-6-chloro-2-[2-(methylamino)-2-oxoethyl]-N-(5-methylisoquinolin-4-yl)-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-2-[2-(methylamino)-2-oxoethyl]-N-(5-methylisoquinolin-4-yl)-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 72.20% | 15.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 47.5 | - | - | - | |||||
| 7gmg | 1.81 Å | 2023-11-08 | X-ray | RJO | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.11% | 23.82% | 7.80% | (4S)-6-chloro-N-(6-fluoroisoquinolin-4-yl)-2-[2-(methylamino)-2-oxoethyl]-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-(6-fluoroisoquinolin-4-yl)-2-[2-(methylamino)-2-oxoethyl]-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 64.50% | 11.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 46.6 | - | - | - | |||||
| 7gmh | 1.85 Å | 2023-11-08 | X-ray | RJX | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.12% | 23.57% | 6.10% | 4-[2-(3-chlorophenyl)acetamido]isoquinoline-6-carboxylic acid | 4-[2-(3-chlorophenyl)acetamido]isoquinoline-6-carboxylic acid | XDS | 67.00% | 14.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 49.3 | - | - | - | |||||
| 7gmi | 2.01 Å | 2023-11-08 | X-ray | RK6 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.82% | 24.53% | 11.60% | 4-[2-(3-chlorophenyl)acetamido]-N-methylisoquinoline-7-carboxamide | 4-[2-(3-chlorophenyl)acetamido]-N-methylisoquinoline-7-carboxamide | XDS | 61.10% | 10.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 39.8 | - | - | - | |||||
| 7gmj | 1.84 Å | 2023-11-08 | X-ray | RKC | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.08% | 23.70% | 6.60% | (3'R)-6-chloro-1'-(isoquinolin-4-yl)-2,3-dihydrospiro[[1]benzopyran-4,3'-pyrrolidin]-2'-one | (3'R)-6-chloro-1'-(isoquinolin-4-yl)-2,3-dihydrospiro[[1]benzopyran-4,3'-pyrrolidin]-2'-one | XDS | 77.30% | 13.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 47.7 | - | - | - | |||||
| 7gmk | 1.84 Å | 2023-11-08 | X-ray | RKR | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.52% | 24.02% | 5.80% | 2-(3-chlorophenyl)-N-[6-(dimethylamino)isoquinolin-4-yl]acetamide | 2-(3-chlorophenyl)-N-[6-(dimethylamino)isoquinolin-4-yl]acetamide | XDS | 83.00% | 14.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 44.6 | - | - | - | |||||
| 7gml | 1.91 Å | 2023-11-08 | X-ray | RL0 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.32% | 24.36% | 9.30% | 2-(3-chlorophenyl)-N-{6-[(methanesulfonyl)(methyl)amino]isoquinolin-4-yl}acetamide | 2-(3-chlorophenyl)-N-{6-[(methanesulfonyl)(methyl)amino]isoquinolin-4-yl}acetamide | XDS | 69.70% | 11.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 41.5 | - | - | - | |||||
| 7gmm | 1.84 Å | 2023-11-08 | X-ray | RL8 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.25% | 23.78% | 6.10% | 4-[2-(3-chlorophenyl)acetamido]-N-methylisoquinoline-6-carboxamide | 4-[2-(3-chlorophenyl)acetamido]-N-methylisoquinoline-6-carboxamide | XDS | 72.80% | 14.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 47.2 | - | - | - | |||||
| 7gmn | 2.19 Å | 2023-11-08 | X-ray | RLH | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.46% | 26.78% | 12.90% | 2-(3-chlorophenyl)-N-{7-[2-(pyrrolidin-1-yl)ethoxy]isoquinolin-4-yl}acetamide | 2-(3-chlorophenyl)-N-{7-[2-(pyrrolidin-1-yl)ethoxy]isoquinolin-4-yl}acetamide | XDS | 57.70% | 9.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 21.1 | - | - | - | |||||
| 7gmo | 1.85 Å | 2023-11-08 | X-ray | RLR | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.98% | 24.51% | 8.00% | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[(3R)-2-oxopyrrolidin-3-yl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[(3R)-2-oxopyrrolidin-3-yl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 61.70% | 11.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 39.9 | - | - | - | |||||
| 7gmp | 2.08 Å | 2023-11-08 | X-ray | RM3 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.01% | 25.93% | 15.00% | 2-(3-chlorophenyl)-N-[7-(dimethylamino)isoquinolin-4-yl]acetamide | 2-(3-chlorophenyl)-N-[7-(dimethylamino)isoquinolin-4-yl]acetamide | XDS | 55.70% | 9.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 27.1 | - | - | - | |||||
| 7gmq | 1.86 Å | 2023-11-08 | X-ray | RMI | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.99% | 24.43% | 6.70% | 2-(3-chlorophenyl)-N-{7-[2-(dimethylamino)ethoxy]isoquinolin-4-yl}acetamide | 2-(3-chlorophenyl)-N-{7-[2-(dimethylamino)ethoxy]isoquinolin-4-yl}acetamide | XDS | 70.30% | 13.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 40.8 | - | - | - | |||||
| 7gmr | 1.84 Å | 2023-11-08 | X-ray | RN0 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.36% | 24.94% | 11.70% | 4-[2-(3-chlorophenyl)acetamido]isoquinolin-6-yl methanesulfonate | 4-[2-(3-chlorophenyl)acetamido]isoquinolin-6-yl methanesulfonate | XDS | 68.60% | 9.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 35.7 | - | - | - | |||||
| 7gms | 1.90 Å | 2023-11-08 | X-ray | RNI | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.82% | 25.67% | 12.60% | (4S)-6-chloro-2-[(1-cyanocyclopropyl)methanesulfonyl]-N-{7-[(methanesulfonyl)amino]isoquinolin-4-yl}-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-2-[(1-cyanocyclopropyl)methanesulfonyl]-N-{7-[(methanesulfonyl)amino]isoquinolin-4-yl}-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 64.80% | 8.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 29.4 | - | - | - | |||||
| 7gmt | 1.74 Å | 2023-11-08 | X-ray | ROZ | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.60% | 24.67% | 10.20% | 4-[2-(3-chlorophenyl)acetamido]isoquinoline-6-carboxamide | 4-[2-(3-chlorophenyl)acetamido]isoquinoline-6-carboxamide | XDS | 59.50% | 10.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 38.5 | - | - | - | |||||
| 7gmu | 1.74 Å | 2023-11-08 | X-ray | RPK | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.80% | 25.93% | 9.90% | N-(6-acetamidoisoquinolin-4-yl)-2-(3-chlorophenyl)acetamide | N-(6-acetamidoisoquinolin-4-yl)-2-(3-chlorophenyl)acetamide | XDS | 59.30% | 10.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 27.1 | - | - | - | |||||
| 7gmv | 1.90 Å | 2023-11-08 | X-ray | RPZ | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.39% | 25.19% | 13.90% | 2-[(3'S)-6-chloro-1'-(isoquinolin-4-yl)-2'-oxo-1H-spiro[isoquinoline-4,3'-pyrrolidin]-2(3H)-yl]-N-methylacetamide | 2-[(3'S)-6-chloro-1'-(isoquinolin-4-yl)-2'-oxo-1H-spiro[isoquinoline-4,3'-pyrrolidin]-2(3H)-yl]-N-methylacetamide | XDS | 56.40% | 9.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 33.6 | - | - | - | |||||
| 7gmw | 1.83 Å | 2023-11-08 | X-ray | RQ6 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.64% | 25.55% | 15.60% | (4S)-6-chloro-N-(7-fluoroisoquinolin-4-yl)-2-[2-(methylamino)-2-oxoethyl]-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-(7-fluoroisoquinolin-4-yl)-2-[2-(methylamino)-2-oxoethyl]-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 56.10% | 8.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 30.4 | - | - | - | |||||
| 7gmx | 2.03 Å | 2023-11-08 | X-ray | RQO | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.84% | 26.77% | 16.50% | 4-[2-(3-chlorophenyl)acetamido]-N,N-dimethylisoquinoline-6-carboxamide | 4-[2-(3-chlorophenyl)acetamido]-N,N-dimethylisoquinoline-6-carboxamide | XDS | 57.00% | 7.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 21.1 | - | - | - | |||||
| 7gmy | 1.84 Å | 2023-11-08 | X-ray | RQF | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.09% | 25.53% | 12.50% | (4S)-6-chloro-N-(7-fluoroisoquinolin-4-yl)-1-oxo-2-[2-oxo-2-(propylamino)ethyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-(7-fluoroisoquinolin-4-yl)-1-oxo-2-[2-oxo-2-(propylamino)ethyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 61.10% | 9.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 30.5 | - | - | - | |||||
| 7gmz | 1.62 Å | 2023-11-08 | X-ray | RR0 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.97% | 24.79% | 9.60% | (2r,4r)-6-chloro-N-(7-fluoroisoquinolin-4-yl)-2,3-dihydro-4H-2,4-methano-1-benzopyran-4-carboxamide | (2r,4r)-6-chloro-N-(7-fluoroisoquinolin-4-yl)-2,3-dihydro-4H-2,4-methano-1-benzopyran-4-carboxamide | XDS | 56.60% | 10.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 37.4 | - | - | - | |||||
| 7gn0 | 1.81 Å | 2023-11-08 | X-ray | RRD | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.50% | 24.73% | 12.60% | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-{2-[(oxetan-3-yl)amino]-2-oxoethyl}-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-{2-[(oxetan-3-yl)amino]-2-oxoethyl}-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 52.10% | 5.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 37.8 | - | - | - | |||||
| 7gn1 | 1.90 Å | 2023-11-08 | X-ray | RRU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.97% | 26.08% | 14.80% | 1-{[(3'S)-6-chloro-1'-(isoquinolin-4-yl)-2'-oxo-1H-spiro[isoquinoline-4,3'-pyrrolidine]-2(3H)-sulfonyl]methyl}cyclopropane-1-carbonitrile | 1-{[(3'S)-6-chloro-1'-(isoquinolin-4-yl)-2'-oxo-1H-spiro[isoquinoline-4,3'-pyrrolidine]-2(3H)-sulfonyl]methyl}cyclopropane-1-carbonitrile | XDS | 62.70% | 8.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 26.1 | - | - | - | |||||
| 7gn2 | 1.74 Å | 2023-11-08 | X-ray | RS6 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.57% | 24.16% | 14.30% | (4S)-6-chloro-N-(7-fluoroisoquinolin-4-yl)-1-oxo-2-{2-oxo-2-[(propan-2-yl)amino]ethyl}-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-(7-fluoroisoquinolin-4-yl)-1-oxo-2-{2-oxo-2-[(propan-2-yl)amino]ethyl}-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 62.90% | 8.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 43.4 | - | - | - | |||||
| 7gn3 | 2.01 Å | 2023-11-08 | X-ray | RSL | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.08% | 26.13% | 18.70% | 4-[2-(3-chlorophenyl)acetamido]isoquinolin-7-yl methanesulfonate | 4-[2-(3-chlorophenyl)acetamido]isoquinolin-7-yl methanesulfonate | XDS | 61.30% | 7.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 25.6 | - | - | - | |||||
| 7gn4 | 1.72 Å | 2023-11-08 | X-ray | RT4 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.76% | 24.10% | 13.10% | (4S)-6-chloro-2-[2-(cyclopropylamino)-2-oxoethyl]-N-(isoquinolin-4-yl)-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-2-[2-(cyclopropylamino)-2-oxoethyl]-N-(isoquinolin-4-yl)-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 64.00% | 9.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 43.8 | - | - | - | |||||
| 7gn5 | 1.86 Å | 2023-11-08 | X-ray | RT9 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.61% | 25.44% | 21.00% | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[(1-methoxycyclopropyl)methanesulfonyl]-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[(1-methoxycyclopropyl)methanesulfonyl]-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 61.90% | 7.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 31.3 | - | - | - | |||||
| 7gn6 | 2.44 Å | 2023-11-08 | X-ray | RTS | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.03% | 27.44% | 34.10% | (4S)-6-chloro-N-(isoquinolin-4-yl)-4-methyl-2-[2-(methylamino)-2-oxoethyl]-1,1-dioxo-1,2,3,4-tetrahydro-1lambda~6~,2-benzothiazine-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-4-methyl-2-[2-(methylamino)-2-oxoethyl]-1,1-dioxo-1,2,3,4-tetrahydro-1lambda~6~,2-benzothiazine-4-carboxamide | XDS | 65.00% | 4.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 17.0 | - | - | - | |||||
| 7gn7 | 2.35 Å | 2023-11-08 | X-ray | RV0 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 19.47% | 26.44% | 24.40% | (4S)-6-chloro-4-ethyl-N-(isoquinolin-4-yl)-1,1-dioxo-1,2,3,4-tetrahydro-1lambda~6~,2-benzothiazine-4-carboxamide | (4S)-6-chloro-4-ethyl-N-(isoquinolin-4-yl)-1,1-dioxo-1,2,3,4-tetrahydro-1lambda~6~,2-benzothiazine-4-carboxamide | XDS | 51.40% | 4.3 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 23.4 | - | - | - | |||||
| 7gn8 | 1.90 Å | 2023-11-08 | X-ray | RPZ | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.04% | 25.02% | 13.80% | 2-[(3'S)-6-chloro-1'-(isoquinolin-4-yl)-2'-oxo-1H-spiro[isoquinoline-4,3'-pyrrolidin]-2(3H)-yl]-N-methylacetamide | 2-[(3'S)-6-chloro-1'-(isoquinolin-4-yl)-2'-oxo-1H-spiro[isoquinoline-4,3'-pyrrolidin]-2(3H)-yl]-N-methylacetamide | XDS | 69.80% | 8.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 34.9 | - | - | - | |||||
| 7gn9 | 2.15 Å | 2023-11-08 | X-ray | RVL | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.27% | 25.32% | 24.00% | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[1-(methylcarbamoyl)cyclopropyl]-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[1-(methylcarbamoyl)cyclopropyl]-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 58.50% | 6.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 32.3 | - | - | - | |||||
| 7gna | 2.09 Å | 2023-11-08 | X-ray | RVR | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.46% | 26.58% | 24.70% | (4S)-6-chloro-1,1-dioxo-N-(5,6,7,8-tetrahydroisoquinolin-4-yl)-1,2,3,4-tetrahydro-1lambda~6~,2-benzothiazine-4-carboxamide | (4S)-6-chloro-1,1-dioxo-N-(5,6,7,8-tetrahydroisoquinolin-4-yl)-1,2,3,4-tetrahydro-1lambda~6~,2-benzothiazine-4-carboxamide | XDS | 51.80% | 5.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 22.5 | - | - | - | |||||
| 7gnb | 1.67 Å | 2023-11-08 | X-ray | RW0 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.95% | 23.86% | 9.60% | (3R,4S)-6-chloro-N-[7-(methanesulfonyl)isoquinolin-4-yl]-3-methyl-2-[2-(methylamino)-2-oxoethyl]-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (3R,4S)-6-chloro-N-[7-(methanesulfonyl)isoquinolin-4-yl]-3-methyl-2-[2-(methylamino)-2-oxoethyl]-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 58.70% | 11.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 46.3 | - | - | - | |||||
| 7gnc | 1.68 Å | 2023-11-08 | X-ray | RW9 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.95% | 23.84% | 15.90% | 2-[(3'S)-6-chloro-1'-(isoquinolin-4-yl)-1,2'-dioxo-1H-spiro[isoquinoline-4,3'-pyrrolidin]-2(3H)-yl]-N-methylacetamide | 2-[(3'S)-6-chloro-1'-(isoquinolin-4-yl)-1,2'-dioxo-1H-spiro[isoquinoline-4,3'-pyrrolidin]-2(3H)-yl]-N-methylacetamide | XDS | 61.10% | 8.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 46.4 | - | - | - | |||||
| 7gnd | 1.66 Å | 2023-11-08 | X-ray | RVR | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.42% | 23.83% | 11.90% | (4S)-6-chloro-1,1-dioxo-N-(5,6,7,8-tetrahydroisoquinolin-4-yl)-1,2,3,4-tetrahydro-1lambda~6~,2-benzothiazine-4-carboxamide | (4S)-6-chloro-1,1-dioxo-N-(5,6,7,8-tetrahydroisoquinolin-4-yl)-1,2,3,4-tetrahydro-1lambda~6~,2-benzothiazine-4-carboxamide | XDS | 68.90% | 9.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 46.5 | - | - | - | |||||
| 7gne | 1.74 Å | 2023-11-08 | X-ray | RWO | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.71% | 25.07% | 13.30% | (4R)-6-chloro-4-methyl-1,1-dioxo-N-(5,6,7,8-tetrahydroisoquinolin-4-yl)-1,2,3,4-tetrahydro-1lambda~6~,2-benzothiazine-4-carboxamide | (4R)-6-chloro-4-methyl-1,1-dioxo-N-(5,6,7,8-tetrahydroisoquinolin-4-yl)-1,2,3,4-tetrahydro-1lambda~6~,2-benzothiazine-4-carboxamide | XDS | 56.60% | 8.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 34.6 | - | - | - | |||||
| 7gnf | 1.69 Å | 2023-11-08 | X-ray | RWT | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.45% | 25.10% | 12.20% | (4S)-6-chloro-4-methyl-1,1-dioxo-N-(5,6,7,8-tetrahydroisoquinolin-4-yl)-1,2,3,4-tetrahydro-1lambda~6~,2-benzothiazine-4-carboxamide | (4S)-6-chloro-4-methyl-1,1-dioxo-N-(5,6,7,8-tetrahydroisoquinolin-4-yl)-1,2,3,4-tetrahydro-1lambda~6~,2-benzothiazine-4-carboxamide | XDS | 57.90% | 7.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 34.2 | - | - | - | |||||
| 7gng | 1.77 Å | 2023-11-08 | X-ray | RXU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.71% | 24.96% | 11.00% | 2-[(3'S)-6-chloro-2'-oxo-1'-(5,6,7,8-tetrahydroisoquinolin-4-yl)-1H-spiro[isoquinoline-4,3'-pyrrolidin]-2(3H)-yl]-N-methylacetamide | 2-[(3'S)-6-chloro-2'-oxo-1'-(5,6,7,8-tetrahydroisoquinolin-4-yl)-1H-spiro[isoquinoline-4,3'-pyrrolidin]-2(3H)-yl]-N-methylacetamide | XDS | 59.10% | 10.0 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 35.6 | - | - | - | |||||
| 7gnh | 1.78 Å | 2023-11-08 | X-ray | RYB | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.73% | 24.23% | 10.60% | 1-{[(3'S)-6-chloro-1'-{6-[2-(dimethylamino)ethoxy]isoquinolin-4-yl}-2'-oxo-1H-spiro[isoquinoline-4,3'-pyrrolidine]-2(3H)-sulfonyl]methyl}cyclopropane-1-carbonitrile | 1-{[(3'S)-6-chloro-1'-{6-[2-(dimethylamino)ethoxy]isoquinolin-4-yl}-2'-oxo-1H-spiro[isoquinoline-4,3'-pyrrolidine]-2(3H)-sulfonyl]methyl}cyclopropane-1-carbonitrile | XDS | 74.00% | 9.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 42.7 | - | - | - | |||||
| 7gni | 1.64 Å | 2023-11-08 | X-ray | RZF | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.44% | 24.06% | 13.70% | 2-[(3'S)-6-chloro-1'-(6-chloroisoquinolin-4-yl)-1,2'-dioxo-1H-spiro[isoquinoline-4,3'-pyrrolidin]-2(3H)-yl]-N-methylacetamide | 2-[(3'S)-6-chloro-1'-(6-chloroisoquinolin-4-yl)-1,2'-dioxo-1H-spiro[isoquinoline-4,3'-pyrrolidin]-2(3H)-yl]-N-methylacetamide | XDS | 60.70% | 9.1 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 44.3 | - | - | - | |||||
| 7gnj | 1.45 Å | 2023-11-08 | X-ray | RZU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.51% | 22.92% | 10.30% | 1-[(3'S)-6-chloro-1'-(isoquinolin-4-yl)-1,2'-dioxo-1H-spiro[isoquinoline-4,3'-pyrrolidin]-2(3H)-yl]-N-methylcyclopropane-1-carboxamide | 1-[(3'S)-6-chloro-1'-(isoquinolin-4-yl)-1,2'-dioxo-1H-spiro[isoquinoline-4,3'-pyrrolidin]-2(3H)-yl]-N-methylcyclopropane-1-carboxamide | XDS | 100.00% | 10.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 55.8 | - | - | - | |||||
| 7gnk | 1.72 Å | 2023-11-08 | X-ray | S0X | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.22% | 25.26% | 18.80% | 2-[(3'S)-6-chloro-1'-(isoquinolin-4-yl)-2'-oxo-1H-spiro[isoquinoline-4,3'-pyrrolidin]-2(3H)-yl]-N-(cyclopropylmethyl)acetamide | 2-[(3'S)-6-chloro-1'-(isoquinolin-4-yl)-2'-oxo-1H-spiro[isoquinoline-4,3'-pyrrolidin]-2(3H)-yl]-N-(cyclopropylmethyl)acetamide | XDS | 98.20% | 7.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 32.9 | - | - | - | |||||
| 7gnl | 1.68 Å | 2023-11-08 | X-ray | S1U | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 21.78% | 23.93% | 12.80% | (4S)-6-chloro-2-{2-[4-(4-ethylpiperazin-1-yl)anilino]-2-oxoethyl}-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-2-{2-[4-(4-ethylpiperazin-1-yl)anilino]-2-oxoethyl}-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 100.00% | 9.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 45.5 | - | - | - | |||||
| 7gnm | 1.75 Å | 2023-11-08 | X-ray | S1L, NA | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.64% | 25.37% | 14.90% | SODIUM ION; (4S)-6-chloro-N-(isoquinolin-4-yl)-2-{2-[3-(morpholin-4-yl)anilino]-2-oxoethyl}-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-{2-[3-(morpholin-4-yl)anilino]-2-oxoethyl}-1,2,3,4-tetrahydroisoquinoline-4-carboxamide; SODIUM ION | XDS | 100.00% | 9.2 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 32.0 | - | - | - | |||||
| 7gnn | 1.81 Å | 2023-11-08 | X-ray | S39 | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.34% | 25.18% | 17.10% | (4S)-2-{2-[(1,3-benzothiazol-5-yl)amino]-2-oxoethyl}-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-2-{2-[(1,3-benzothiazol-5-yl)amino]-2-oxoethyl}-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 98.30% | 7.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 33.7 | - | - | - | |||||
| 7gno | 1.66 Å | 2023-11-08 | X-ray | S3X | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.92% | 23.23% | 11.50% | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-(2-{[(1S)-1-(4-nitrophenyl)ethyl]amino}-2-oxoethyl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-(2-{[(1S)-1-(4-nitrophenyl)ethyl]amino}-2-oxoethyl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 99.80% | 10.6 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 52.7 | - | - | - | |||||
| 7gnp | 1.55 Å | 2023-11-08 | X-ray | NA, S4X | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.15% | 26.00% | 15.30% | (4S)-6-chloro-2-(2-{[(1r,3R,5R,7S)-3-hydroxyadamantan-1-yl]amino}-2-oxoethyl)-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide; SODIUM ION | SODIUM ION; (4S)-6-chloro-2-(2-{[(1r,3R,5R,7S)-3-hydroxyadamantan-1-yl]amino}-2-oxoethyl)-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 100.00% | 6.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 26.5 | - | - | - | |||||
| 7gnq | 1.53 Å | 2023-11-08 | X-ray | S5L | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 20.44% | 22.35% | 13.40% | (4S)-2-[2-(4-acetamidoanilino)-2-oxoethyl]-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | (4S)-2-[2-(4-acetamidoanilino)-2-oxoethyl]-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide | XDS | 99.90% | 8.8 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 61.7 | - | - | - | |||||
| 7gnr | 1.82 Å | 2023-11-08 | X-ray | RZU | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 23.00% | 25.71% | 24.90% | 1-[(3'S)-6-chloro-1'-(isoquinolin-4-yl)-1,2'-dioxo-1H-spiro[isoquinoline-4,3'-pyrrolidin]-2(3H)-yl]-N-methylcyclopropane-1-carboxamide | 1-[(3'S)-6-chloro-1'-(isoquinolin-4-yl)-1,2'-dioxo-1H-spiro[isoquinoline-4,3'-pyrrolidin]-2(3H)-yl]-N-methylcyclopropane-1-carboxamide | XDS | 100.00% | 7.4 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 28.9 | - | - | - | |||||
| 7gns | 1.62 Å | 2023-11-08 | X-ray | S6K | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 24.69% | 26.50% | 13.80% | 1-{[(3'S,4'R)-6-chloro-1'-(isoquinolin-4-yl)-4'-methyl-2'-oxo-1H-spiro[isoquinoline-4,3'-pyrrolidine]-2(3H)-sulfonyl]methyl}cyclopropane-1-carbonitrile | 1-{[(3'S,4'R)-6-chloro-1'-(isoquinolin-4-yl)-4'-methyl-2'-oxo-1H-spiro[isoquinoline-4,3'-pyrrolidine]-2(3H)-sulfonyl]methyl}cyclopropane-1-carbonitrile | XDS | 99.70% | 8.5 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 22.9 | - | - | - | |||||
| 7gnt | 1.56 Å | 2023-11-08 | X-ray | S7C | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.81% | 25.10% | 12.80% | 1-{[(3'S,4'R)-6-chloro-4'-ethyl-1'-(isoquinolin-4-yl)-2'-oxo-1H-spiro[isoquinoline-4,3'-pyrrolidine]-2(3H)-sulfonyl]methyl}cyclopropane-1-carbonitrile | 1-{[(3'S,4'R)-6-chloro-4'-ethyl-1'-(isoquinolin-4-yl)-2'-oxo-1H-spiro[isoquinoline-4,3'-pyrrolidine]-2(3H)-sulfonyl]methyl}cyclopropane-1-carbonitrile | XDS | 98.50% | 8.9 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 34.2 | - | - | - | |||||
| 7gnu | 1.48 Å | 2023-11-08 | X-ray | S7U | SARS-CoV-2 | NSP5 (3CLpro) | PanDDA fragment screening | 22.95% | 24.51% | 10.60% | 1-{[(3'S)-6-chloro-1'-(isoquinolin-4-yl)-2'-oxo-1H-spiro[isoquinoline-4,3'-piperidine]-2(3H)-sulfonyl]methyl}cyclopropane-1-carbonitrile | 1-{[(3'S)-6-chloro-1'-(isoquinolin-4-yl)-2'-oxo-1H-spiro[isoquinoline-4,3'-piperidine]-2(3H)-sulfonyl]methyl}cyclopropane-1-carbonitrile | XDS | 100.00% | 9.7 | Diamond (I04-1) | BUSTER | 2023-08-11 | - | - | - | - | - | N/A | - | - | 3C-like proteinase | - | - | No | - | - | 39.9 | - | - | - | |||||
| 8bfo | 1.99 Å | 2023-11-08 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 21.20% | 28.59% | N/A | - | XDS | 98.60% | 7.75 | ALBA (XALOC) | REFMAC | 2022-10-26 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 11.6 | - | - | - | |||||||
| 8bfq | 1.86 Å | 2023-11-08 | X-ray | - | SARS-CoV-2 | NSP5 (3CLpro) | 19.22% | 25.34% | N/A | - | XDS | 97.10% | 10.06 | ALBA (XALOC) | REFMAC | 2022-10-26 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 32.2 | - | - | - | |||||||
| 8bg1 | 2.88 Å | 2023-11-08 | X-ray | NAG | SARS-CoV-2 | Spike | 20.21% | 24.44% | 13.65% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 96.95% | 9.38 | SLS (X06SA) | PHENIX | 2022-10-27 | - | - | - | - | - | N/A | - | - | pT1511 Fab heavy chain; pT1511 Fab light chain; Spike protein S1 | - | - | No | - | - | 40.7 | - | - | - | ||||||
| 8bg2 | 2.10 Å | 2023-11-08 | X-ray | NAG | SARS-CoV-2 | Spike | 18.90% | 22.73% | 9.49% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 94.11% | 9.19 | SLS (X06SA) | PHENIX | 2022-10-27 | - | - | - | - | - | N/A | - | - | Spike protein S1; pT1375 single-chain Fv | - | - | No | - | - | 57.8 | - | - | - | ||||||
| 8bg3 | 1.90 Å | 2023-11-08 | X-ray | NAG | SARS-CoV-2 | Spike | 18.48% | 21.40% | 9.14% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 92.35% | 8.46 | SLS (X06SA) | PHENIX | 2022-10-27 | - | - | - | - | - | N/A | - | - | Spike protein S1; pT1610 single-chain Fv | - | - | No | - | - | 70.6 | - | - | - | ||||||
| 8bg4 | 1.60 Å | 2023-11-08 | X-ray | NAG | SARS-CoV-2 | Spike | 19.45% | 20.77% | 9.06% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.90% | 21.14 | SLS (X06SA) | PHENIX | 2022-10-27 | - | - | - | - | - | N/A | - | - | Spike protein S1; pT1611 single-chain Fv | - | - | No | - | - | 76.0 | - | - | - | ||||||
| 8bg5 | 2.05 Å | 2023-11-08 | X-ray | NAG | SARS-CoV-2 | Spike | 19.63% | 21.66% | 11.30% | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 99.75% | 16.31 | SOLEIL (PROXIMA 1) | PHENIX | 2022-10-27 | - | - | - | - | - | N/A | - | - | pT1631 single-chain Fv; Spike protein S1 | - | - | No | - | - | 68.4 | - | - | - | |||||||
| 8bg6 | 4.11 Å | 2023-11-08 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX; phenix.real_space_refine | 2022-10-27 | - | - | - | - | - | N/A | - | - | pT1644 Fab light chain; Spike glycoprotein; pT1644 Fab heavy chain | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 8bg8 | 3.64 Å | 2023-11-08 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2022-10-27 | - | - | - | - | - | N/A | - | - | Spike glycoprotein; pT1696 Fab light chain; pT1696 Fab heavy chain | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||||
| 8bga | 1.98 Å | 2023-11-08 | X-ray | QQL | SARS-CoV-2 | NSP5 (3CLpro) | 19.27% | 23.72% | N/A | 4-methoxy-~{N}-[(2~{S})-4-methyl-1-[[(2~{S})-4-nitro-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-pentan-2-yl]-1~{H}-indole-2-carboxamide | 4-methoxy-~{N}-[(2~{S})-4-methyl-1-[[(2~{S})-4-nitro-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-pentan-2-yl]-1~{H}-indole-2-carboxamide | XDS | 98.70% | 9.13 | ESRF (ID30B) | REFMAC | 2022-10-27 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 47.6 | - | - | - | ||||||
| 8bgd | 1.62 Å | 2023-11-08 | X-ray | QH0 | SARS-CoV-2 | NSP5 (3CLpro) | 19.14% | 24.39% | N/A | (phenylmethyl) ~{N}-[(2~{S})-4-methyl-1-[[(2~{S})-4-nitro-1-[(3~{R})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-pentan-2-yl]carbamate | (phenylmethyl) ~{N}-[(2~{S})-4-methyl-1-[[(2~{S})-4-nitro-1-[(3~{R})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-pentan-2-yl]carbamate | XDS | 99.40% | 21.94 | ALBA (XALOC) | REFMAC | 2022-10-27 | - | - | - | - | - | N/A | - | - | 3C-like proteinase nsp5 | - | - | No | - | - | 41.3 | - | - | - | ||||||
| 8hes | 2.20 Å | 2023-11-08 | X-ray | NAG | SARS-CoV-2 | Spike | 19.62% | 24.08% | 15.20% | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | XDS | 98.15% | 7.8 | SLS (X06SA) | PHENIX | 2022-11-08 | 37452031 | - | - | - | - | N/A | - | - | NIV-10 Fab L-chain; NIV-10 Fab H-chain; Spike protein S1 | - | - | No | - | 44.2 | - | - | - | |||||||
| 7tpi | 2.30 Å | 2023-11-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2022-01-25 | - | - | - | - | - | N/A | - | - | Spike glycoprotein - Ectodomain | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 8f4p | 3.70 Å | 2023-11-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | PHENIX | 2022-11-11 | - | - | - | - | - | N/A | - | - | Anti-S1 Nanobody; Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | |||||||
| 8ix3 | 3.98 Å | 2023-11-15 | Cryo-EM | - | SARS-CoV-2 | Spike | N/A | N/A | N/A | - | - | - | () | 2023-03-31 | 37855619 | - | - | - | - | N/A | - | - | light chain of 1G11; BA.4/5 variant spike protein; heavy chain of 1G11 | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 8u1t | - | 2023-11-15 | NMR | - | SARS-CoV-2 | Envelope | N/A | N/A | N/A | - | - | - | () | 2023-09-02 | 37914906 | - | - | - | - | N/A | - | - | Envelope small membrane protein | N/A | - | No | - | N/A | N/A | N/A | - | ||||||||||
| 8u28 | 3.10 Å | 2023-11-15 | Cryo-EM | NAG | SARS-CoV-2 | Spike | N/A | N/A | N/A | 2-acetamido-2-deoxy-beta-D-glucopyranose | 2-acetamido-2-deoxy-beta-D-glucopyranose | - | - | () | 2023-09-05 | - | - | - | - | - | N/A | - | - | Spike glycoprotein | N/A | - | No | - | - | N/A | N/A | N/A | - | ||||||||
| 8urb | 3.40 Å | 2023-11-15 | Cryo-EM | - | PEDV | NSP7/NSP8/NSP12 | N/A | N/A | N/A | RNA (55-MER); RNA (33-MER) | - | - | - | () | 2023-10-25 | 36993498 | ZN | - | - | - | N/A | - | - | RNA (55-MER); RNA (33-MER); nsp7; nsp8; nsp12 | N/A | - | No | - | N/A | N/A | N/A | RNA (55-MER); RNA (33-MER) | - |
Other resources
- https://github.com/thorn-lab/coronavirus_structural_task_force
Andrea Thorn’s group at University of Wurzburg - http://kinemage.biochem.duke.edu/
Jane and David Richardson’s group at Duke University - https://www.globalphasing.com/buster/wiki/index.cgi?Covid19
Gerard Bricogne’s group at Global Phasing - https://zhanglab.ccmb.med.umich.edu/COVID-19/
Yang Zhang’s group at University of Michigan - https://coronavirus3d.org/
Adam Godzik’s group at University of Riverside - https://isolde.cimr.cam.ac.uk/what-isolde/
Tistan Croll group at University of Cambridge - https://www.rcsb.org/news?year=2020&article=5e74d55d2d410731e9944f52&feature=true
Varius links to COVID-19/SARS-CoV-2 Resources from RCSB PDB - https://www.ebi.ac.uk/pdbe/covid-19
Protein Data Bank in Europe COVID-19 website - https://www.covid19dataportal.org/
The European Bioinformatics Institute (EMBL) COVID-19 data portal - https://proteindiffraction.org/search/?q=COVID-19
SARS-CoV-2 raw diffraction data at the Integrated Resource for Reproducibility in Structural Biology
Funding
This work was supported in part by NIGMS grant R01-GM132595 and the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. We acknowledge support of the National Science Center (NCN, Poland), grant No. 2020/01/0/NZ1/00134. The project was also partly funded by the Polish National Agency for Academic Exchange under Grant No. PPN/BEK/2018/1/00058/U/00001 and by the FWF (Austrian Science Foundation) grant P 32821.
Attributions
Full list of used libraries, fonts, and images is available at the Attributions page.
Citing and contact
If you find this server useful, please cite: Wlodawer et al. (2020) Ligand-centered assessment of SARS-CoV-2 drug target models in the Protein Data Bank, FEBS Journal 287, 3703-3718. or Brzezinski et al. (2021) Covid‐19.bioreproducibility.org: A web resource for SARS‐CoV‐2‐related structural models, Protein Science 30, 115-124. .
The PQ1 metric was calculated according to Brzezinski et al. (2020) On the evolution of the quality of macromolecular models in the PDB, FEBS Journal 287(13), 2685-2698. Structure alignment to the reference cell was done using ACHESYM: Kowiel et al. (2014) ACHESYM: an algorithm and server for standardized placement of macromolecular models in the unit cell, Acta Cryst. D70, 3290-3298.
The server is under constant development and updated regularly. If you find a bug, have a comment, or want to collaborate, contact us: , , .
Copyright © 2020 All rights reserved